Abstract
LEAFY is a floral meristem identity gene in Arabidopsis thaliana, which plays a key role during flowering. BrcLEAFY (BrcLFY), a FLO/LFY homologue, was cloned from Pak Choi by reverse transcription-polymerase chain reaction (RT-PCR), and its expression patterns in different organs and the apex at different developmental stages, as well as during vernalization, were analysed by quantitative real-time PCR (qPCR). The full cDNA of BrcLFY was 1353 bp in length with an open reading frame of 1260 bp encoding 419 amino acids. qPCR results showed that BrcLFY expression gradually increased during the vegetative growth stage and increased sharply at flower differentiation stage 1, with the highest expression level at flower differentiation stage 5. When comparing BrcLFY expression in different organs, we concluded that the level in young and mature leaves was significantly higher than in the apex and other tissues at flower differentiation stage 3 and visible flower bud stage and fully-bloom stage. The BrcLFY expression in germinated seeds and seedlings with five leaves was analysed during cold treatment for 0 d, 10 d and 20 d at 4 ºC and the results showed that BrcLFY gene expression rose gradually with prolonged low temperature.
Introduction
Pak Choi (Brassica rapa subsp. chinensis), one of the typical vegetables in China, is not only cultivated all over the country, but has also been recently introduced into other countries in Asia, North America and Europe. Pak Choi is a typical seed-vernalization-responsive vegetable, which requires a prolonged cold treatment combined with the onset of long days, ensuring that flowering occurs in the spring. However, the overwintering period and temperatures between varieties differs markedly. Plants with a weak vernalization requirement undergo early bolting in the spring, whereas those with longer vernalization and lower temperature requirement, flower with difficulty, resulting in low seeds yield and quality.[Citation1] Therefore, research on the flowering mechanism in Pak Choi has important practical and theoretical value.
Flowering has been extensively studied in the model plant Arabidopsis, where it is regulated by multiple pathways.[Citation2] These pathways converge at the flower meristem identity genes, such as LEAFY (LFY), APETALA1 (AP1) and CAULIFLOWER (CAL), as described by Komeda [Citation3] and switch on their expression.[Citation4–7] LFY plays a pivotal role in a plant's flowering, because its expression precedes other meristem identity genes and activates floral organ identity genes,[Citation8–10] and it is also responsible for the initial steps in flower differentiation.
LFY has two roles in flower development of Arabidopsis: firstly it allows the transition from vegetative to reproductive stage as a pathway integrator and secondly, it is responsible for the initiation of floral primordium as a meristem identity gene and activates floral organ identity genes.[Citation3,Citation11,Citation12] Since the first LFY homologous gene, FLO from Antirrhinum, was isolated by Coen et al.,[Citation13] LFY family genes have been isolated successively in many plant species, including rice,[Citation14] wheat,[Citation15] pea,[Citation16] tomato,[Citation17] etc. The identified LFY family genes are highly conserved both in their structure and functions.[Citation18,Citation19] However, the expression patterns are different. A large number of reports suggested that there are differences in the expression patterns of LFY family genes in different species.[Citation20] RFL, the LFY homologue in rice, is expressed only in panicles, but not in leaves or flowers.[Citation14] In strawberries FaLFY was mainly transcribed in the reproductive tissues, especially in flower buds, but no transcription was detected in the vegetative tissues, neither in the four whorls of the flower organs.[Citation21] Unlike rice and strawberry, AcLFY was detected during the whole period of bolting and flowering in onion, and it had a high transcription level in the inflorescence meristem at the beginning of bolting, but almost no transcription in floral organs. After the development of floral organs, the AcLFY gene was mainly expressed in leaves.[Citation22] DFL, the LFY homologue gene from chrysanthemum, is expressed also in stems, leaf primordial tissues and developing inflorescence bracts,[Citation23] and is dramatically upregulated in induced shoot apical meristems.[Citation24] FA, the tomato ortholog of LFY, is expressed in both vegetative and floral meristems, in leaf primordia and leaves, as well as in the four floral organs.[Citation17]
In the present work, BrcLEAFY (BrcLFY) gene was isolated from the shoot apex of Pak Choi variety ‘75#’ by reverse transcription-polymerase chain reaction (RT-PCR). The expression patterns in different organs and the apex at different developmental stages, as well as during vernalization, were analysed by quantitative real-time PCR (qPCR) so as to determine the BrcLFY gene expression pattern during the transition from vegetative growth to reproductive growth. This will help to reveal the relationship between gene expression and floral induction and clarify LFY's role during flowering, which might further improve the understanding of the plant flowering molecular mechanism.
Materials and methods
Plant material
Pak Choi variety '75#' was an easy bolting inbred line and was a kind gift from the Institute of Vegetables Research, Shanxi Academy of Agricultural Sciences.
Seeds were soaked in 55 ºC water for 10 min and kept for 2 h at room temperature. Then, they were placed in petri dishes with double filter paper on the bottom and kept in growth chamber at 25 ºC with 16 h/day light until the radicle broke through the seed coat. Thereafter, they were placed in the refrigerator at 4 ºC for 20 d in dark. The filter paper was kept moist during the period to ensure seed germination.[Citation25] After treatment, the seedlings were planted in plastic nutrition pots filled with sphagnum peat / vermiculite substrate mixture (3:1, v/v), and grown in the greenhouse under 30 °C daytime maximum temperature and 10 ºC night minimum temperature with approximately 11 h of natural light. Standard agricultural practices were applied throughout.
Paraffin sectioning and scanning electron microscopy
After seven leaves were unfolded, the shoot apices were peeled and observed by a stereo microscope (Olympus SZX16) every 2 d, so as to determine the developmental stages. When the tissue in specific developmental stage was found, it was fixed, sectioned and dewaxed, according to the modified protocol, described by Garcês and Sinha.[Citation26,Citation27] The tissue was stained as follows: 70% (v/v) ethanol for 5 min, 1% (v/v) safranin solution for 2 h, 70% (v/v) ethanol for 1 min, 85% (v/v) ethanol for 1 min, 100% ethanol for 2 min, xylene for 3 min and xylene for another 3 min. At the end the slides were mounted in neutral balsam, observed and photographed using Olympus IX81 microscope. Some samples were fixed using the same methods, dried using liquid CO2 in a Bio-Rad 750 critical-point dryer, mounted on scanning electron microscope (SEM) stubs and coated by gold pounder. Images were obtained from the SEM (JSM-6490LV, JEOL Japan) at 15 kV.
Total RNA extraction and cDNA synthesis
Young inflorescences with flower buds (0.1 g) were collected from the plants and used for total RNA extraction. Total RNA was extracted using QIAGEN RNeasy Plant Mini Kit with DNase I (QIAGEN, Germany), according to the suggested protocol. The integrity of total RNA was verified by running the samples on a 1% agarose gel. First-strand cDNA was synthesized in two steps by using PrimeScript® 1st Strand cDNA Synthesis Kit (TaKaRa, D6110S, Japan) according to the manufacturer's instructions. Firstly, 10 μL mixture containing 1.0 μg total RNA, 1 μL 10 mmol/L dNTP mix and 1 μL 50 µmol/L Oligo dT primer, was incubated at 65 °C for 5 min and then placed on ice for at least 1 min. Meanwhile, a 10 μL mixture including 4 µL 5 × PrimerScript Buffer, 0.5 µL RNase inhitor (40 U/μL), 1 µL Primer Script Rtase (200 U/μL) and 4.5 µL RNase free dH2O, was made and added to the reaction solution. After that the mixture was incubated at 42 °C for 50 min and terminated at 95 °C for 5 min. The obtained cDNA was kept in –20 °C until further use.
Gene cloning and sequencing
Forward primer (5′-AGAAAGAGAATATGGATCCTGAAGG-3′) and reverse primer (5′-CCAGCCAAAACGAGTAAAAACTAGAGCC-3′) were designed using Primer 3 software based on Brassica rapa subsp. pekinensis clone KBrH005L20 in GenBank (accession number: AC232542.1). The homologue of LFY was amplified by standard RT-PCR in the following reaction conditions: 5 min at 94 °C; 10 cycles of 15 s at 94 °C, 20 s at 60–51 °C and 2 min at 72 °C; 40 cycles of 15 s at 94 °C, 20 s at 55 °C, 2 min at 72 °C; followed by a 5 min extension at 72 °C. The PCR products were analysed on 1% agarose gel and cloned into pMD19-T Vector (TaKaRa, Japan), according to the manufacturer's instructions. At least two replicates were sequenced in Beijing BGI Biotechnology Ltd. (Beijing, P. R. China).
Bioinformatics analysis
The sequences were analysed using Basic Local Alignment Search Tool (BLAST) (http://www.ncbi.nlm.nih.gov/BLAST) in National Center of Biotechnology Information (NCBI). For phylogenetic and sequence alignment analysis, LFY-like protein sequences from other plants were obtained from the GenBank Database (www.ncbi.nlm.nih.gov/genbank). The accession numbers of the genes used in this study are as follows: LFY (A. thaliana, NM_125579), ZFL1 (Zea mays, AY179882), RFL (Oryza sativa, AB005620), CFL (Cucumis sativus, AF059320), LFYa (Brassica rapa, HQ615937), RsLFY1 (Raphanus sativus, AB449248), AFL2 (Malus × domestica, AB162034), FaLFY (Fragaria nanassa, JN788260), FAL (Lycopersicon esculentum, AF197934), FLO (Antirrhinum majus, M55525), BOFH (Brassica oleracea, Z18362), AcLFY (Allium cepa, JX275962), NLF (Narcissus tazetta, DQ979379), GinNdly (Ginkgo biloba, AF105111), NEEDLY (Pinus radiata, U76757), UNI (Pisum sativum AF010190). Multiple protein sequence alignment was performed using DNAMAN version 6.0 (Lynnon Biosoft Corporation, USA) (www.lynon.com). A phylogenetic tree for the putative relatives of BrcLFY was reconstructed with the MEGA 5.10. The nearest neighbour-joining method was applied to create the tree. Bootstrap values were derived from 1,000 replicates and the scale bar (0.05) indicates a 5% change of amino acids.
Gene expression analysis
To study the BrcLFY expression pattern, we assessed its relative expression levels in different organs including young leaves, mature leaves and roots and in the shoot apex at vegetative and reproductive stages by qPCR. In addition, germinated seeds (seed vernalization) and the seedlings with five leaves (seedling vernalization) were treated for 0 d, 10 d and 20 d at 4 ºC to investigate BrcLFY gene expression during vernalization. The samples in every stage were collected, frozen quickly in liquid nitrogen and kept at –80 °C. Total RNA was extracted using RNA Simple Total RNA Kit (Tiangen, DP432, P. R. China) with DNase I, following the manufacturer's instructions. cDNA synthesis was performed from 500 ng total RNA following the procedure of 15 min at 37 °C and 5 s at 85 °C using PrimeScript® RT Reagent Kit for Perfect Real Time (TaKaRa, DRR037A, Japan).
Forward and reverse primers for BrcLFY were designed with the on-line Primer 3 software. The primers were: 5′-GGCAGGCAAAGATGAAGAAG-3′ and 5′-CCACGGTCTTTAGCAATGGT-3′. ACTIN gene of Pak Choi was used as a reference gene and the forward and reverse primers were 5′-GTTGCTATCCAGGCTGTTCT-3′ and 5′-AGCGTGAGGAAGAGCATAAC-3′, respectively. qPCR was performed by using SYBR® Premix Ex TaqTM (Tli RNaseH Plus) (TaKaRa, DRR820A, Japan). Each 25 μL reaction contained 12.5 μL of SYBR Premix Ex Tag, 1 μL of each of the forward and reverse primers, 8.5 μL of deionized water (dH2O), 2 μL of template cDNA. The thermal cycling conditions were an initial denaturation step for 30 s at 95 °C, followed by 40 PCR cycles of 5 s at 95 °C, 15 s at 55 ºC and 20 s at 72 ºC. The relative levels of gene expression were calculated using the comparative 2−ΔCT method.[Citation28] Each RNA sample was from six apexes of the plants and the data consisted of three biological replicates and two technical replicates.
Results and discussion
Flower differentiation in Pak Choi
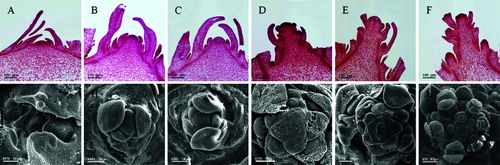
Based on the flower differentiation classification in Brassica rapa subsp. pekinensis [Citation29] and Arabidopsis,[Citation30] the Pak Choi flower development process was divided into six stages, based on the paraffin sectioning and the scanning electron microscopy (): stage 0 (vegetative meristem): the shoot meristem was flat and surrounded by a few conical leaf primordia (A) and 1(G)); stage 1 (vegetative meristem transit to reproductive meristem): shoot apical meristem started to swell and elongate and became dome-shaped. Leaf primordia continued to differentiate in the periphery of the shoot apex. Stage 1 was observed to be the period of transition from vegetative to reproductive development (B) and 1(H)); stage 2 (early flower primordia): the shoot apex meristem continued to expand and elongate and became surrounded by 2–3 round bumps, i.e. flower primordia, leaf primordia differentiation was terminated (C) and 1(I)); stage 3 (floral differentiation): round bumps around the apical meristem increased continuously, forming clusters (D) and 1(J)); stage 4 (pedicel elongation): pedicels began to elongate, but did not became too long, sepals were undifferentiated, clusters of round bumps differentiated as primary structures of flower buds (E) and 1(K)); stage 5 (individual flower differentiation): pedicels continued to elongate and primordia of sepal, pistil and stamen began to form (F) and 1(L)).
Cloning and sequence analysis of BrcLFY gene
A full-length cDNA of LFY-like gene was cloned from the apex of Pak Choi by RT-PCR, the gene was designated as BrcLFY. The PCR product contained 1353 bp with an open reading frame for BrcLFY of 1,260 bp. BrcLFY (GenBank Accession No. KJ924980) was predicted to encode a putative protein of 419 amino acids with predicted molecular weight of 46.32 kDa and isoelectric point (pI) of 6.55.
An alignment of predicted amino acid sequence with other reported LFY-like genes revealed that the deduced amino acid sequence of BrcLFY showed high identity to other plant LFY-like genes and shared 50.9% identity to Z. mays (ZFL1), 51.3% identity to O. sativa (RFL), 69% identity to C. sativus (CFL), 89% identity to A. thaliana (LFY), 92% identity to R. sativus (RsLFY1) and 99% identity to B. rapa (LFYa) ().
Figure 2. Multiple alignments of amino acid sequence of BrcLFY with LFY homologues from other species.

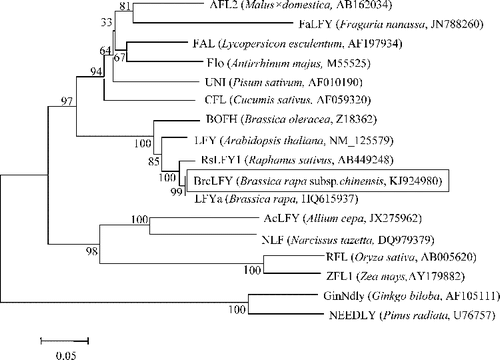
To determine the phylogenetic relationship between BrcLFY and other LFY-like proteins, MEGA5.10 software was used to construct a phylogenetic tree with some predicted amino acid sequences from different species. All of the sequences from the same taxa were clustered together. The BrcLFY protein was more closely related to dicotyledon FLO/LFY-like proteins than monocotyledon or gymnosperm homologues. All the sequences from Brassicaceae species were classified into one cluster. The BrcLFY was more closely related to the LFY-like proteins from cruciferous vegetables, especially Brassica vegetables (). This conclusion was consistent with the result from Suchandra et al. [Citation7] that further verified a closer genetic relationship between them.
BrcLFY gene expression in the process of flowering transition
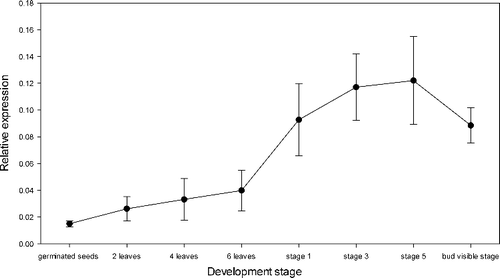
BrcLFY expression was detected in different developmental stages of the shoot apical meristem from the vegetative growth stage to visible flower bud stage (). The expression level was the lowest in seedlings with cotyledons only. The expression level increased slowly with plant growth. A dramatic elevation was observed at flower differentiation stage 1. The expression continued to increase during the flower differentiation process and was the highest at stage 5, after which the BrcLFY gene expression in the apex decreased rapidly, after the flower differentiation was completed. The gene expression trend of BrcLFY was consistent with the expression of LFY gene in Arabidopsis, where LFY was extensively expressed during the vegetative phase. During flowering its expression was strong in floral anlagen and in stage 1 and stage 2 flowers, and persisted at high levels throughout floral primordia until stage 3, when expression abated in the centre of the flower.[Citation9,Citation31] These results suggested that BrcLFY plays a similar role to its homologue during floral buds differentiation and acts as an integrator, regulating flowering time and determining meristem identity in Pak Choi.[Citation3,Citation32]
Figure 4. BrcLFY gene expression in the apex during different developmental stages. Note: error bars represent the standard deviations based on three replicates
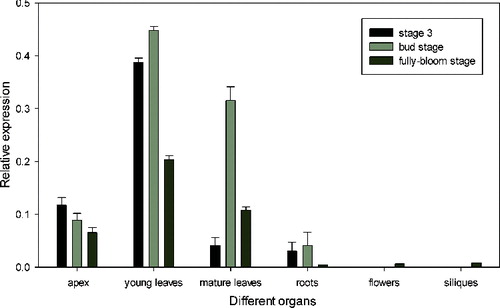
The organ-specific expression patterns of BrcLFY at three developmental stages were investigated based on the qPCR expression profile (). BrcLFY transcription could be detected in all tissues, but was the most highly expressed in young leaves, moderately expressed in mature leaves and weakly expressed in the roots in all three stages. These results were different from the results, obtained with Arabidopsis, in which the LFY gene expression in young and mature leaves was significantly higher than that in shoot apical meristem and other tissues during the vegetative growth stage, but once the plant grew to the reproductive developmental stage, the flower primordium had higher LFY RNA level than the leaf primordium.[Citation7] On the other hand, the obtained results were similar to the expression pattern observed in Brassica juncea,[Citation7] where the expression of BjLFY in mature leaves was the highest, followed by the stem and bracts. However, LFY homologues from some plants (FLO gene from snapdragon, FaLFY gene from strawberry, BOFH gene from cauliflower and AFL1 gene from apple) have not been reported to be expressed in the leaves.[Citation13,Citation21,Citation33,Citation34] This variation in the pattern and expression levels of this gene in different species may indicate the existence of a functional divergence in the FLO/LFY homologues. Many reports [Citation35,Citation36] have indicated that LFY and its homologues are multi-functional genes, because they can be expressed not only in the meristem. LFY homologues could possibly be involved in a variety of morphogenetic stages and other developmental tissues, such as leaf formation and silique development.[Citation16,Citation37–39] Functional studies of the role of BrcLFY in Pak Choi should be further demonstrated.
BrcLFY gene expression in response to low temperature
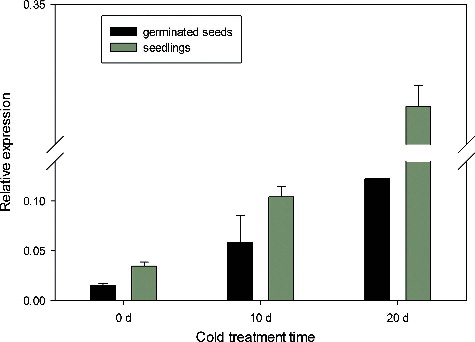
Germinated seeds (seed vernalization) and the seedlings with five leaves (seedling vernalization) were treated for 0 d, 10 d and 20 d at 4 ºC to investigate BrcLFY gene expression during vernalization. BrcLFY gene expression showed an increasing trend with prolonged cold treatment in both types of plants. The gene expression levels in the seedlings were higher than that in the germinated seeds during the period (). However, the plants remained vegetative and did not initiate floral differentiation (seed vernalization results shown in , seedlings vernalization data not shown), which was similar to the results from tobacco and Narcissus tazetta.[Citation40,Citation41] In tobacco, the expression of LFY-like was up-regulated at low temperature.[Citation40] Stored at low temperature, N. tazetta remained morphologically vegetative and did not produce flowers. However, LFY homologous gene expression was significantly higher than in other storage conditions.[Citation41] Because the BrcLFY expression level increased during vernalizaion, the plant remained in vegetative growth. This indicated that the flower differentiation was not solely dependent on increasing the BrcLFY expression. Therefore, the intermediate links and signalling mechanisms need further analyses.
Conclusions
Pak Choi is an important and typical seed-vernalization-responsive vegetable. LFY plays a key role during plant flowering. In this study, BrcLFY, a LFY homologue, was cloned from Pak Choi and phylogenetic, and sequence alignment was performed; the expression patterns in different organs and the apex at different developmental stages, as well as during vernalization, were analysed by qPCR. The results showed that BrcLFY played a role both in vegetative and reproductive development, promoting transition from vegetative growth to reproductive development in Pak Choi. Moreover, BrcLFY responded to vernalization. These results will help to establish conventional and molecular breeding systems in Pak Choi and facilitate the research on the flowering mechanisms in the plant.
Acknowledgements
The authors would like to thank Prof Yuan-huai Han and Prof Donald Grierson for their critical revision of this manuscript and thank Dr Xiao-yun Jia for submitting the sequence.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Liu RY, Hou LP, Wang L, et al. Effect analysis of low temperature promoting flowering in Chinese cabbage. ACTA Agriculturae Boreali-Sinica. 2009;24(6):193–197.
- Putterill J, Laurie R, Macknight R. It's time to flower: the genetic control of flowering time. Bioessays. 2004;26(4):363–373.
- Komeda Y. Genetic regulation of time to flower in Arabidopsis thaliana. Annu Rev Plant Biol. 2004;55(1):521–535.
- Bowman JL, Alvarez J, Weigel D, et al. Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development. 1993;11:721–743.
- Busch MA, Bomblies K, Weigel D. Activation of a floral homeotic gene in Arabidopsis. Science. 1999;285(5427):585–587.
- Gustafson-Brown C, Savidge B, Yanofsky MF. Regulation of the Arabidopsis floral homeotic gene APETALA1. Cell. 1994;76(1):131–143.
- Suchandra DR, Mukesh S, Neera BS. Cloning and expression studies of a LFY cDNA from Brassica juncea. Adv Biosci Biotechnol. 2011;2(4):248–254.
- Alejandra Mandel MA, Gustafson-Brown C, Savidge B, et al. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature. 1992;360(6401):273–277.
- Weigel D, Alvarez J, Smyth DR, et al. LEAFY controls floral meristem identity in Arabidopsis. Cell. 1992;69(5):843–859.
- Ingram GC, Goodrich J. Wilkinson MD, et al. Parallels between UNUSUAL FLORAL ORGANS and FIMBRIATA, genes controlling flower development in Arabidopsis and Antirrhinum. Plant Cell. 1995;7(9):1501–1510.
- Madueño F, Ruiz-Garcia L, Wilkinson M, et al. Different roles of flowering time genes in the activation of floral initiation genes in Arabidopsis. Int J Dev Biol. 1996;40:S125–S126.
- Hamès C, Ptchelkine D, Grimm C, et al. Structural basis for LEAFY floral switch function and similarity with helix-turn-helix proteins. EMBO J. 2008;27(19):2628–2637.
- Coen ES, Romero JM, Doyle S, et al. Floricaula: a homeotic gene required for flower development in Antirrhinum majus. Cell. 1990;63(6):1311–1322.
- Kyozuka J, Konishi S, Nemoto K, et al. Down-regulation of RFL, the FLO/LFY homolog of rice, accompanied with panicle branch initiation. P Natl Acad Sci USA. 1998;95(5):1979–1982.
- Shitsukawa N, Takagishi A, Ikari C, et al. WFL, a wheat FLORICAULA/LEAFY ortholog, is associated with spikelet formation as lateral branch of the inflorescence meristem. Genes Genet Syst. 2006;81(1):13–20.
- Hofer J, Turner L, Hellens R, et al. UNIFOLIATA regulates leaf and flower morphogenesis in pea. Curr Biol. 1997;7(8):581–587.
- Molinero-Rosales N, Jamilena M, Zurita S, et al. FALSIFLORA, the tomato orthologue of FLORICAULA and LEAFY, controls flowering time and floral meristem identity. Plant J. 1999;20(6):685–693.
- Hou LP, Li ML, Li YL. The basis of molecular biology in plants flowering. Journal of Shanxi Agricultural Sciences. 2002;30(2):82–88.
- Lv LL, Sun GM, Liu YG, et al. Progress of LEAFY homologous gene study. Acta Botanica Boreali-Occidentalia Sinica. 2011;31(1):0197–0203.
- Sliwinski MK, White MA, Maizel A, et al. Evolutionary divergence of LFY function in the mustards Arabidopsis thaliana and Leavenworthia crassa. Plant Mol Biol. 2006;62(1–2):279–289.
- Liu YX, Zou DM, Li H, et al. Isolation and expression analysis of LFY homologue from strawberry. Acta Horticulturae Sinica. 2012;39(5):861–868.
- Ye YY, Chen D, Wang Y. Cloning and expression analysis of flowering related gene AcLFY from Allium cepa. Acta Horticulturae Sinica. 2013;40(2):283–291.
- Ma YP, Fang XH, Chen F, et al. DFL, a FLORICAULA/LEAFY homologue gene from Dendranthema lavandulifolium is expressed both in the vegetative and reproductive tissues. Plant Cell Rep. 2008;27(4):647–654.
- Ma YP, Zhou YZ, Wang YZ, et al. CnFL, a FLORICAULA/LEAFY homolog in Chrysanthemum nankingense is dramatically upregulated in induced shoot apical meristems. Biochem Syst Ecol. 2013;50:114–120.
- Fu C, Song HX, Hou LP, et al. Effects of different vernalization days on flowering of different varieties in Pak Choi. Highlights of Science Paper Online. 2014;7(19):1957–1962.
- Garcês H, Sinha N. Fixing and sectioning tissue from the plant Kalanchoë daigremontiana. Cold Spring Harb Protoc. 2009a;10:pdb. prot5301.
- Garcês H, Sinha N. In situ hybridization in the plant Kalanchoë daigremontiana. Cold Spring Harb Protoc. 2009b;10:pdb. prot 5302.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408.
- Li SX, Li SJ. Brassica campestris L.ssp.chinensis flower bud differentiation and leaf head formation. Scientia Agricultura Sinica. 1964;6:36–40.
- Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2(8):755–767.
- Blázquez MA, Soowal LN, Lee I, et al. LEAFY expression and flower initiation in Arabidopsis. Development. 1997;124:3835–3844.
- Jack T. Molecular and genetic mechanisms of floral control. Plant Cell. 2004;16(suppl_1):S1.
- Anthony RG, James PE, Jordan BR. Cloning and sequence analysis of a FLO/LFY homologue isolated from cauliflower (Brassica oleracea L. var. botrytis). Plant Mol Biol.1993;22(6):1163–1166.
- Wada M, Cao QF, Kotoda N, et al. Apple has two orthologues of FLORICAULA/LEAFY involved in flowering. Plant Mol Biol. 2002;49(6):567–577.
- Moyroud E, Kusters E, Monniaux M, et al. LEAFY blossoms. Trends Plant Sci. 2010;15(6):346–352.
- Neta R, David-Schwartz R, Peretz Y, et al. Flower development in garlic: the ups and downs of gaLFY expression. Planta. 2011;233(5):1063–1072.
- Meng QC, Zhang CH, Huang F, et al. Molecular cloning and characterization of a LEAFY-like gene highly expressed in developing soybean seeds. Seed Sci Res. 2007;17(4):297–302.
- Oshima S, Nomura K. RsLFY, a LEAFY homologue gene in radish (Raphanus sativus), is continuously expressed in vegetative, reproductive and seed development. Plant Biotechnol J. 2008;25(6):579–582.
- Wang HL, Chen JH, Wen JQ, et al. Control of compound leaf development by FLORICAULA/LEAFY ortholog SINGLE LEAFLET1 in Medicago truncatula. Plant Physiol. 2008;146(4):1759–1772.
- Shi YC. Identification of signal molecules involved in early flowering Promoted by low temperature and signal transduction Path way exploration [dissertation]. Zhengzhou (CHN): Henan Agricultural University; 2010.
- Noy-Porat T, Kamenetsky R, Eshel A, et al. Temporal and spatial expression patterns of the LEAFY homologue NLF during florogenesis in Narcissus tazetta. Plant Sci. 2010;178(2):105–113.