ABSTRACT
Plants can be used as biological indicators in assessing the damage done by bioaccumulation of heavy metals and their negative impact on the environment. In the present research, Roman nettle (Urtica pilulifera L.) was employed as a bioindicator for cadmium (Cd) pollution. The comparisons between unexposed and exposed plant samples revealed inhibition of the root growth (∼25.96% and ∼45.92% after treatment with 100 and 200 µmol/L Cd concentrations, respectively), reduction in the total soluble protein quantities (∼53.92% and ∼66.29% after treatment with 100 and 200 µmol/L Cd concentrations, respectively) and a gradual genomic instability when the Cd concentrations were increased. The results indicated that alterations in randomly amplified polymorphic DNA (RAPD) profiles, following the Cd treatments, included normal band losses and emergence of new bands, when compared to the controls. Also, the obtained data from F1 plants, utilized for analysis of genotoxicity, revealed that DNA alterations, occurring in parent plants due to Cd pollution, were transmitted to the next generation.
Introduction
Cadmium (Cd) is one of the most toxic elements which increases its accumulation in natural and/or agricultural soils via anthropogenic activities.[Citation1,Citation2] Moreover, the soluble capacity of Cd increases its effects on the contaminated soils.[Citation3] The bioaccumulation of Cd into all organisms and into the food chain could pose a direct threat to the ecosystems.[Citation4]
Cd is uptaken by the plants’ root systems and causes many defects at different levels of the entire plant.[Citation5] The presence of Cd in plants influences the water balance and brings out many stress symptoms, such as reduction in photosynthesis and disturbances in the mineral nutrient uptake by affecting the plasma membranes' permeability,[Citation6] inhibits the gas exchange, respiration,[Citation6] cell proliferation,[Citation7] carbohydrate metabolism [Citation8] and causes further serious problems, which may result even in death.[Citation9] The toxic levels of Cd may cause damages in different components of the cells, including membranes, proteins and DNA.[Citation9–11] More particularly, the changes involve modifications of genetic constitutions of the plants, including abnormalities in chromosomes,[Citation9] nucleolar structures [Citation12] and mitotic index.[Citation12] The generation of reactive oxygen species can occur during oxidative stress in cells due to heavy metal toxicity at a biochemical level and may cause lipid peroxidation, enzyme inactivation [Citation13] and DNA damage.[Citation14] The accumulation of oxidized proteins and lipid peroxides, induced by Cd exposure, was observed in pea.[Citation15]
Toxic effects of Cd have been documented in many plants, such as Pisum sativum,[Citation16] Allium cepa,[Citation17] Hordeum vulgare,[Citation18] Arabidopsis thaliana,[Citation19] Glycine max,[Citation20] Vicia faba,[Citation21] Zea mays,[Citation22] Nicotiana tabacum,[Citation23] Allium sativum,[Citation21] Thlaspi caerulescens,[Citation24] Silene vulgaris,[Citation25] Salix sp. [Citation26] and Tradescantia sp.[Citation27]
Randomly amplified polymorphic DNA-polymerase chain reaction (RAPD-PCR) is used for DNA analysis in the field of genotoxicity, as a sensitive method, capable of detecting variations in genome profiles, like normal band losses or appearances of new bands, following toxic exposures.[Citation28,Citation29] The purpose of the present work was to make assessments of the DNA damage, done by Cd, by using the RAPD-PCR method, and to compare the RAPD profiles of control and 100 and 200 µmol/L Cd exposed Parents and F1 plantlets of Urtica pilulifera. This is one of the important traditional and widely used plants in Europe and Turkey, because of its pharmaceutical properties. It is also a weed for cultivated land and waste areas. It is common for the coastal parts of Turkey, such as Canakkale, Istanbul, Samsun and Izmir.[Citation30] Furthermore, this study could be used to ascertain whether genotoxic effects of Cd are inherited or not by the next generation.
Materials and methods
In vitro development of U. pilulifera plantlets
U. pilulifera seeds (Parents) were surface sterilized for 1 min with 50% (v/v) ethyl alcohol and for 5 min with deionized water (Millipore). After the sterilization, they were germinated on sterilized compost in small vessels for a two-week period and then young plantlets, in which shoot lengths reached about 3–4 cm, were transferred into standard plastic pots. During a two-month growing period, the plantlets were kept in a growth chamber under fluorescent tubes, giving an irradiance of 5000 lx (16 h/8 h day/night photoperiod), a temperature of 23 ± 2 °C and a relative humidity of 45%–50%. The stress applications to the experimental groups of replicates (control (without application of Cd stress) parental and F1 (from seeds of parent plants) groups, each including eight samples and parental and F1 groups treated with 100 and 200 μmol/L Cd concentrations grown from the seeds of parent plants treated with 100 and 200 μmol/L Cd concentrations, each including eight samples) were conducted by using 40 mL spiked Hoagland's nutrient solutions [Citation31] containing 0 µmol/L (control), 100 µmol/L and 200 µmol/L Cd (in the form of CdCl2∙H2O) at two-day intervals.
Determination of growth inhibition rates of U. pilulifera
After two months of incubation, the root and stem length (cm) of U. pilulifera were measured and the parents’ root growth inhibition rates (RGIR) and stem growth inhibition rates (SGIR) (%) were determined. The reproduction of U. pilulifera was carried out by self-pollination. During the experimental phase, the self-pollination practices were performed in order to have seeds for the production of F1 plantlets for each group to understand whether the DNA alterations occurring in the parent plants, due to Cd pollution, are going to be transmitted to the next generation ().
Table 1. Seed types and experimental groups.
The seeds of the parent plants were harvested after two months of stress treatment. After the surface-sterilization of the plants with 1% (v/v) sodium hypochlorite for 10 min and deionized water, they were grown in soil with test solutions (40 mL spiked Hoagland's nutrient solutions containing 100 and 200 µmol/L Cd in the form of CdCl2·H2O) and one control (distilled water). Nine different experimental groups of F1 plants (from parent plants’ seeds) were prepared. Their parents were treated with 100 or 200 μmol/L Cd concentrations (). Stock Cd (CdCl2∙2H2O) solutions (1000 μmol/L) were diluted into 100 or 200 μmol/L concentrations with distilled water (). In total, 25 seeds were used for each test solution of plant groups (control groups from parental and F1 plants (from parent plants’ seeds) and 100 and 200 μmol/L Cd-treated groups from parental and F1 plants (from parent plants’ seeds treated with 100 and 200 μmol/L Cd concentrations)). After adding the test solution, the plants were incubated in a climatic conditioner at 23 °C in the dark for 7 d.
Determination of total protein in roots of U. pilulifera
The content of total soluble protein (TSP) in the root tips of U. pilulifera (µg/g FW) was measured by spectroscopic techniques (PG instruments T60), according to Bradford.[Citation32]
Detection of genotoxic alterations in U. pilulifera
DNA was isolated by using the DNeasy Plant DNA Extraction Mini Kit (Qiagen), according to the manufacturer's instructions. Estimations of RAPD band profiles were based on comparisons done by using a standard sample (Gene Ruler™ 100 bp DNA Ladder, ready-to-use, Thermo-Fermentas) in a 2% agarose gel, stained with ethidium bromide and visualized under UV-light. The results were documented using Owl EasyCast B2 mini gel electrophoresis system (Thermo Scientific). During the generation of amplification products, each PCR mixture was set up as follows: 1 x PCR buffer (NH4)2SO4 (pH 8.8), 0.2 mmol/L from each deoxyribonucleoside triphosphate (dNTP) (2 mmol/L dNTP mix), 25 pmol of primer OPA08 5'-GTGACGTAGG-3' from QIAGEN Operon RAPD® 10mer Kits, 20 ng–200 ng genomic DNA and 0.5 U of Taq DNA Polymerase. The mixture was filled up with sterile deionized water to a final volume of 25 μL. The PCR chemicals were obtained from Thermo-Fermentas. For the RAPD analysis, 20 random 10-mer primers were used to amplify the genomic DNA extracted from the Cd-treated and untreated U. pilulifera cells (). The primer, designated as OPA08, was used for further steps, because it is capable of producing reproducible and strong bands with the most distinguishable banding profiles between Cd-treated and untreated samples.
Table 2. Sequences and resulting bands of some representing primers used for RAPD amplification of genomic DNA from Cd-treated and control U. pilulifera root cells.
A reaction tube containing all reaction components, except the template DNA, was set up for each reaction as a negative control. The PCR amplifications were programmed for an initial denaturation temperature of 95 °C prior to PCR cycling, to fully denature the template DNA for 3 min, for providing an efficient utilization of template DNA in the first amplification cycle and for preventing poor yields of PCR products. The following configuration was suitable for 45 PCR cycles: a denaturation temperature of 94 °C for 1 min, a primer annealing temperature of 37 °C for 1 min, an elongation temperature of 72 °C for 2 min and a final extension step temperature of 72 °C for 5 min. The amplifications were performed by using a Techne Endurance TC-512 Gradient Thermal Cycler.
DNA variations were identified according to the scored RAPD profiling data, showing normal band losses and occurrences of new bands. The molecular sizes of the amplification products were estimated by using the Gel-Doc 2000 analyzer system (BioRAD) and the Quantity One Program version 4.4.1.
Statistical analyses
Experimental results were statistically analysed by using a variance analysis and Tukey's test. The standard deviation was significant at P < 0.05 and P < 0.001.
Results and discussion
U. pilulifera is an astringent and a galactagogue plant that has antiasthmatic, anti-dandruff, depurative, diuretic, haemostatic and hypoglycemic effects.[Citation30,Citation33] It is a stimulating tonic for medicinal purposes. It is especially used as a remedy for diabetes mellitus, eczema, rheumatism, haemorrhoids, hyperthyroidism, bronchitis and cancer.[Citation30,Citation33]
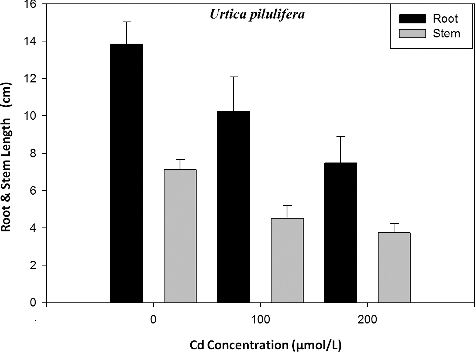
The root and stem growth inhibition rates were analysed in the parent type of U. pilulifera seedlings in response to treatment with different Cd concentrations. The results obtained from the experiments are shown in and . Following the exposure to Cd, the root and stem lengths of U. pilulifera seedlings gradually decreased from 13.83 cm (control) and 7.3 cm (control) to 10.24 cm and 4.8 cm, respectively after treatment with 100 μmol/L Cd and to 7.48 cm and 3.7 cm after treatment with 200 μmol/L Cd, respectively (). After two months of Cd exposure, RGIR and SGIR of plantlets treated with 100 and 200 μmol/L Cd concentration were found to gradually decrease from ∼25.96% and ∼34.25% to ∼45.92% and ∼49.32%, respectively (). The data for the TSP levels are also shown in . The TSP levels in root tips of plantlets were recorded as 334.6 ± 13.4 µg/g FW (P < 0.05), 154.2 ± 7.5 µg/g FW (P < 0.001) and 112.8 ± 6.9 µg/g FW (P < 0.001) at 0, 100 and 200 µmol/L Cd concentrations, respectively. The TSP rates were decreased from ∼53.92% to ∼66.29%. There was a positive correlation between Cd concentration and TSP content in U. pilulifera seedlings, with a correlation coefficient of r = 0.94.
Table 3. Effects of Cd on the root/stem growth inhibition rates and on the amount of total soluble protein in U. pilulifera.
Inhibitory effects of heavy metals on growth and uptake and accumulations of metal elements in Urticaceae members were also shown in previous studies. Such examples are: the altered growth of U. pilulifera upon application of Al stress [Citation34]; reductions on stomatal parameters of U. pilulifera due to excessive aluminium (Al) and Cd exposures [Citation35]; heavy metal (Cd, Cu and Zn) uptake capabilities of Urtica dioica [Citation36]; bioaccumulation of heavy metals by U. dioica.[Citation37–40]
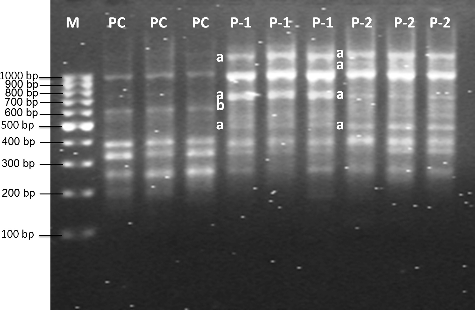
For the RAPD-PCR analysis, 17 out of the 20 random 10-mer primers (85%) produced strong banding patterns, whereas the others (15%) failed to amplify DNA. Totally, 62 amplification products were detected by using 17 primers. Furthermore, the most distinguishable banding profiles (eight strong bands) were produced by the Cd-treated and untreated samples with the primer designated as OPA08. Representative RAPD profiles are shown in and as well as in and. Differentiated Cd-induced RAPD fingerprints were identified by comparisons done between exposed and unexposed (control) samples. These differentiations were indicated as normal band losses and occurrences of new bands.
Figure 3. RAPD profiles of genomic DNA from root tips of F1 type U. pilulifera seedlings exposed to different Cd concentrations.
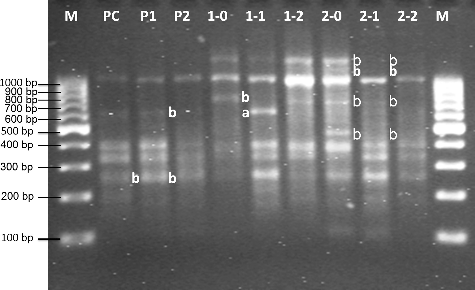
Table 4. Changes of total bands, polymorphic bands and bands in parental type of U. pilulifera seedlings in comparison with parents, 100 µmol/L Cd-treated U. pilulifera, 200 µmol/L Cd-treated U. pilulifera and control.
Table 5. Changes of total bands and polymorphic bands in Cd-treated F1 type of U. pilulifera seedlings in comparison with the control.
Cd-treated and untreated U. pilulifera parent plants are compared in and . F1 plants records are shown in and . The molecular size of the bands obtained with OPA08 ranged from 203 to 1291 bp. Molecular sizes of extra bands, approximately 508, 623, 755, 1082, and 1140 bp, appeared after treatment with 100 and 200 μmol/L Cd concentrations ( and). After treatment with 100 and 200 μmol/L Cd concentrations, 203, 481, 623, 631, 770, 1127, 1291 bp normal RAPD bands disappeared ( and ). When parental control plants (PC), parental plants treated with 100 μmol/L Cd concentration (P1) and parental plants treated with 200 μmol/L Cd concentration (P2) from F1 generation were compared, band alterations were observed ( and ), as seen in the Cd-treated parent plants ( and ). The alterations were loss of some bands (203 and 623 bp) ( and, and). P1 and P2 were created by using untreated parental group from the first generation (). P1 and P2 were then exposed to Cd concentrations of 100 and 200 μmol/ L only for seven days. When P1 and P2 from the second generation and P-1 and P-2 (exposed to Cd concentrations of 100 and 200 μmol/L for two months) from the first generation were compared, DNA damages done by Cd were more severe in P-1 and P-2 seedlings. When PC, 1-0 and 2-0 were compared, disappearance of normal bands (770 and 1291 bp) and appearance of new band (623 bp) were observed ( and ). The groups designated as 1-0 and 2-0 were created by using parental P-1 and P-2 groups (exposed to Cd concentrations of 100 and 200 μmol/L for two months) from first generation. After that, 1-0 and 2-0 were not exposed to Cd stress at any level. When a comparison was done between PC from the second generation and 1-0 and 2-0, the DNA alterations were noticed. This indicates that DNA alterations show a hereditary behaviour. When 1-0, 1-1 and 1-2, and 2-0, 2-1 and 2-2 were compared with each other, once again, alterations on genetic material were seen, such as appearance of new bands (623 bp) or disappearing of normal bands (203, 481, 623, 770, 1127, 1291 bp) ( and ). When a comparison between parental groups and F1 groups was done, DNA alterations in genetic material, seen in F1 groups, were more severe and it was revealed that DNA alterations occurring in parent plants due to Cd pollution, were transmitted to the next generation.
The evaluation of alterations, using the genomic template stability in the genome of U. pilulifera, was done based on the comparisons in RAPD profiles from Cd-treated and untreated samples along with reductions in root and stem growth and soluble protein content of root tips, as well. Following the morphological and physiological parameters, the genomic template stability decreases, as recorded by RAPD-PCR, which also gives some valuable information of Cd-induced hazards to U. pilulifera.
In previous studies, DNA alterations induced by genotoxins were detected in many organisms including a range of plants, aquatic invertebrates and bacterial species using reproducible RAPD profiles.[Citation41] Numerous investigations in the detection of genotoxic effects of heavy metals, can be given as examples. Some of these investigations included organisms such as U. dioica,[Citation42] Evernia prunastri L. Arch.,[Citation43] Rutilus rutilus,[Citation44] Solanum melongena,[Citation45] Zea mays,[Citation46,Citation47] Danio rerio,[Citation48] Arabidopsis thaliana,[Citation49] Egyptian clover and Sudan grass.[Citation50] The present study confirmed that, Cd induces genomic DNA modifications, observed in RAPD profiles as alterations, including normal band losses and occurrences of new bands, when compared with controls. Moreover, there was an inverse relationship between the frequency of band losses and Cd doses applied. It seemed that the number of band losses increased when higher Cd doses were applied ( and , and). Modifications of RAPD patterns were likely due to one or a combination of the following events: (1) alterations that occurred in the oligonucleotide priming sites led to the creation of new sites, accessible to the primers. These sites are used for the amplification and their creation was mainly due to genomic rearrangements, because probabilities of occurrences of mutations in 10-base long primer binding sites are unlikely. On the other hand, genomic rearrangements occur in much longer fragments; (2) interactions between the damaged DNA and DNA Polymerase in U. pilulifera plantlets.[Citation18,Citation51] DNA polymerization in the PCR reaction can be stopped or slowed down due to the emergence of one or a combination of these events.[Citation41]
Cd is shown to induce a variety of DNA damages, including single- and double-strand breaks, modified bases, abasic sites, DNA-protein cross-links, oxidized bases, 8-hydroxyguanine and even bulky adducts in organisms.[Citation52] The kinetics of the PCR events can be affected by the presence of one of the diverse types of DNA damages mentioned above.[Citation53] Mutations, large deletions, and/or homologous recombination are the causes of structural change or changes in DNA sequences, leading to the accessibility of some oligonucleotide priming sites to oligonucleotide primers, resulting in the appearance of new PCR products.[Citation54]
In this study, Cd had inhibitory effect on the growth of root and stem parts, as well as on the TSP content. According to the RAPD profile results, the appearances of new bands occurred after treatment with 100 and 200 μmol/L Cd concentrations. The generation of new bands related to the level of DNA damage, the efficiency of DNA repairs and replications, is the result of genomic template instability.[Citation53] Furthermore, DNA variations in parent and F1 groups showed a hereditary behaviour, where Cd induces growth inhibition and DNA damage in the control, 100 and 200 μmol/L-Cd treated groups. Thus, this study showed that organisms’ characteristics can be affected by environmental factors. These changes are sometimes transmissible to the offspring. Recent researches indicated that environmental changes can cause transmission of similar heritable changes rapidly.[Citation55,Citation56]
The pollutant-induced RAPD profile changes have been used to compare the alterations in genomic DNA template stability and the genotoxic effect with other parameters.[Citation18,Citation41] Cd-induced reduction in root and stem growth inhibition rates and in the TSP level correlated well with the changes in RAPD profiles. This finding showed that the extent of the DNA damage, done by Cd, seemed serious in the majority of cells in the root and stem of U. pilulifera plantlets. At the highest Cd concentration, it seemed that a high level of DNA damage occurred because of the reduced DNA replication.
Conclusions
Gradual reduction in root growth and in the amount of TSPs, and gradual genomic instabilities after treatment with 100 and 200 µmol/L Cd concentrations, were detected by the comparisons done between unexposed and exposed U. pilulifera parent groups. The alterations in RAPD profiles following Cd treatments included normal band losses and emergence of new bands, when compared to the controls. Also, F1 generation plants, utilized for analysis of genotoxicity, produced similar to their parents’ data, revealing that DNA alterations occurring in parent plants due to Cd pollution were transmitted to the next generation. Finally, the present results suggested that using the data from RAPD-PCR analysis, along with the data from physiological parameters, can be a valuable source in assessing Cd toxicity. To demonstrate the validity of the results from the RAPD-PCR analysis, systematic sequencing of genomic targets of Cd toxicity can further be done. Furthermore, DNA damage induced by Cd seems to be transmissible to the offspring.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Yu Z, Zhou Q. Growth responses and cadmium accumulation of Mirabilis jalapa L. under interaction between cadmium and phosphorus. J Hazard Mater. 2009;167:38–43.
- Ci D, Jiang D, Wollenweber B, et al. Genetic variance in cadmium tolerance and accumulation in wheat materials differing in ploidy and genome at seedling stage. J Agron Crop Sci. 2010;196:302–310.
- Daud MK, Sun Y, Dawood M, et al. Cadmium-induced functional and ultrastructural alterations in roots of two transgenic cotton cultivars. J Hazard Mater. 2009;161:463–473.
- Zhang F, Zhang H, Wang G, et al. Cadmium-induced accumulation of hydrogen peroxide in the leaf apoplast of Phaseolus aureus and Vicia sativa and the roles of different antioxidant enzymes. J Hazard Mater. 2009;168:76–84.
- Vitoria AP, Rodriguez APM, Cunha M, et al. Structural changes in radish seedlings (Raphanus sativus) exposed to cadmium. Biol Plant. 2003;47:561–568.
- Seregin IV, Ivanov VB. Physiological aspects of cadmium and lead toxic effects on higher plants. Russ J Plant Physl. 2001;48:523–544.
- Rosas I, Carbajal ME, Gomez-Arroyo S, et al. Cytogenetic effects on cadmium accumulation on water hyacinth (Eichornia crassipes). Environ Res. 1984;33:386–395.
- Moya JL, Ros R, Picazo I. Influence of cadmium and nickel on growth, net photosynthesis and carbohydrate distribution in rice plants. Photosynth Res. 1993;36:75–80.
- Bertin G, Averbeck D. Cadmium: cellular effects, modifications of biomolecules, modulation of DNA repair and genotoxic consequences. Biochimie. 2006;88:1549–1559.
- Zhang HY, Jiang YN, He ZY, et al. Cadmium accumulation and oxidative burst in garlic (Allium sativum). J Plant Physiol. 2005;162:977–984.
- Liu W, Yang YS, Li PJ, et al. Risk assessment of cadmium contaminated soil on plant DNA damage using RAPD and physiological indices. J Hazard Mater. 2009;161:878–883.
- Zhang YX, Yang XL. The toxic effects of cadmium on cell division and chromosomal morphology of Hordeum vulgare. Mutat Res. 1994;312:121–126.
- Sun RL, Zhou QX, Sun FH, et al. Antioxidative defense and praline/phytochelatin accumulation in a newly discovered Cd-hyperaccumulator, Solanum nigrum L. Environ Exp Bot. 2007;60:468–476.
- Gichner T. DNA damage induced by indirect and direct acting mutagens in catalase-deficient transgenic tobacco: cellular and acellular comet assays. Mutat Res. 2003;535:187–193.
- Sandalio LM, Dalurzo HC, Gomez M, et al. Cadmium-induced changes in the growth and oxidative metabolism of pea plants. J Exp Bot. 2001;364:2115–2126.
- Dixit V, Pandey V, Shyam R. Differential antioxidative responses to cadmium in roots and leaves of pea (Pisum sativum L. cv. Azad). J Exp Bot. 2001;52:1101–1109.
- Jiang WS, Liu DH, Li MX. Effects of Cd+2 on the nucleus in root tip cells of Allium cepa. J Environ Sci. 1994;6:382–386.
- Liu W, Li P, Qi X, et al. DNA changes in barley (Hordeum vulgare) seedlings induced by cadmium pollution using RAPD analysis. Chemosphere. 2005;61:158–167.
- Rodriguez-Serrano M, Romero-Puertas MC, Sparkes I, et al. Corrigendum to “Peroxisome dynamics in Arabidopsis plants under oxidative stress induced by cadmium.” Free Radical Bio Med. 2010;48:979.
- Ghorbanli M, Kaveh SH, Sepehr MF. Effects of cadmium and giberellin on growth and photosynthesis of Glycine max. Photosynthetica. 1999;37:627–631.
- Unyayar S, Celik A, Cekic FO, et al. Cadmium-induced genotoxicity, cytotoxicity and lipid peroxidation in Allium sativum and Vica faba. Mutagen. 2006;21:77–81.
- Ju GC, Li XZ, Rauser WE, et al. Influence of cadmium on the production of γ glutamylcysteine peptides and enzymes of nitrogen assimilation in Zea mays seedlings. Physiol Plant. 1997;101:793–799
- Gorinova N, Nedkovska M, Todorovska E, et al. Improved phytoaccumulation of cadmium by genetically modified tobacco plants (Nicotiana tabacum L.). Physiological and biochemical response of the transformants to cadmium toxicity. Environ Pollut. 2007;145:161–170.
- Robinson BH, Leblanc M, Petit D, et al. The potential of Thlaspi caerulescens for phytoremediation of contaminated soils. Plant Soil. 1998;203:47–56.
- Bringezu K, Lichtenberger O, Leopold I, et al. Heavy metal tolerance of Silene vulgaris. J Plant Physiol. 1999;154:536–546.
- Cosio C, Vollenweider P, Keller C. Localization and effects of cadmium in leaves of a cadmium-tolerant willow (Salix viminalis L.) I. Macrolocalization and phytotoxic effects of cadmium. Environ Exp Bot. 2006;58:64–74.
- Steinkellner H, Mun-Sik K, Helma C, et al. Genotoxic effects of heavy metals: comparative investigation with plant bioassays. Environ Mol Mutagen. 1998;31:183–191.
- Uzonur I, Abasiyanik MF, Bostanci B, et al. Re exploring planaria as a model organism for genotoxicity monitoring by an “Improved Random Amplified Polymorphic DNA” approach. Fresen Environ Bull. 2004;13:1420–1426.
- Kekec G, Sakcali MS, Uzonur I. Assesment of genotoxic effects of boron on wheat (Triticum aestivum L.) and bean (Phaseolus vulgaris L.) by using RAPD analysis. Bull Environ Contam Toxicol. 2010;84:759–764.
- Baytop T. Therapy with medicinal plants in Turkey (past and present). Istanbul: Istanbul University; 1999, p. 1–233.
- Hoagland DR, Arnon DI. The water culture method for growing plants without soil. Journal Circular Vol. 347. Berkeley: Agricultural Experimental Station, University of California; 1950.
- Bradford MM. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein dye-binding. Anal Biochem. 1976;72:248–254.
- Kavalali G, Tuncel H, Goksel S, et al. Hypoglycemic activity of Urtica pilulifera in streptozotocin-diabetic rats. J Ethnopharmacol. 2003;84:241–245.
- Dogan I, Ozyigit II, Demir G. Influence of aluminum on mineral nutrient uptake and accumulation in Urtica pilulifera L. J Plant Nutr. 2014;37:469–481.
- Ozyigit II, Akinci S. Effects of some stress factors (aluminum, cadmium and drought) on stomata of Roman Nettle (Urtica pilulifera L.). Notulae Botanicae Horti Agrobotanici Cluj-Napoca. 2009;37(1):108–115.
- Otte ML, Wijte HBM. Environmental variation between habitats and uptake of heavy metals by Urtica dioica. Environ Monitor Assess. 1993;28:263–275.
- Notten MJM, Oosthoek AJP, Rozema J, et al. Heavy metal concentrations in a soil–plant–snail food chain along a terrestrial soil pollution gradient. Environ Pollut. 2005;138(1):178–190.
- Malizia D, Giuliano A, Ortaggi G, et al.. Common plants as alternative analytical tools to monitor heavy metals in soil. Chem Cent J. 2012;(Suppl 2):s6.
- Zeidler M. Heavy metals in two herbs species (River Morava, Czech Republic). Pol J Ecol. 2005;53(2):185–195.
- Maobe MAG, Gatebe E, Gitu L, et al. Profile of heavy metals in selected medicinal plants used for the treatment of diabetes, malaria and pneumonia in Kisii region, Southwest Kenya. Glob J Pharmacol. 2012;6(3):245–251.
- Atienzar FA, Jha NA. The random amplified polymorphic DNA (RAPD) assay and related techniques applied to genotoxicity and carcinogenesis studies: a critical review. Mutat Res. 2006;613:76–102.
- Gjorgieva D, Panovska TK, Ruskovska T, et al. Influence of heavy metal stress on antioxidant status and DNA damage in Urtica dioica. Biomed Res Int. 2013;2013:1–6.
- Cansaran-Duman D, Atakol O, Aras S. Assessment of air pollution genotoxicity by RAPD in Evernia prunastri L. Arc. from around iron-stell factory in Karabuk, Turkey. J Environ Sci. 2011;23(7):1171–1178.
- Salem ZB, Capelli N, Grisey E, et al. First evidence of fish genotoxicity induced by heavy metals from landfill leachates: the advantage of using the RAPD-PCR technique. Ecotoxicol Environ Saf. 2014;101:90–96.
- Korpe DA, Aras S. Evaluation of copper-induced stress on eggplant (Solanum melongena L.) seedlings at the molecular and population levels by use of various biomarkers. Environ Mutagen. 2011;719:29–34.
- Erturk FA, Ay H, Nardemir G, et al. Molecular determination of genotoxic effects of cobalt and nickel on maize (Zea mays L.) by RAPD and protein analyses. Toxicol Ind Health. 2012;29:662–671.
- Erturk FA, Nardemir G, Ay H, et al. Determination of genotoxic effects of boron and zinc on Zea mays using protein and random amplification of polymorphic DNA analyses. Toxicol Ind Health. 2015;31(11):1015–1023.
- Orieux N, Cambier S, Gonzalez P, et al. Genotoxic damages in zebrafish submitted to a polymetallic gradient displayed by the Lot River (France). Ecotoxicol Environ Saf. 2011;74(4):974–983.
- Liu W, Sun L, Zhong M, et al. Cadmium-induced DNA damage and mutatons in Arabidopsis plantlet shoots identified by DNA fingerprinting. Chemosphere. 2012;89(9):1048–1055.
- Aly AA. Application of DNA (RAPD) and ultrastructure to detect the effect of cadmium stress in Egyptian clover Sudan grass plantlets. J Stress Physiol Biochem. 2012;8(1):241–257.
- Rocco L, Valentino IV, Scapigliati G, et al. RAPD-PCR analysis for molecular characterization and genotoxic studies of a new marine fish cell line derived from Dicentrarchus labrax. Cytotechnology. 2014;66(3):383–393.
- Hsiao CJ, Stapleton SR. Characterization of Cd-induced molecular events prior to cellular damage in primary rat hepatocytes in culture: activation of the stress activated signal protein JNK and transcription factor AP-1. J Biochem Mol Toxicol. 2004;18:133–142.
- Bowditch BM, Albright DG, Williams JGK, et al. Use of randomly amplified polymorphic DNA markers in comparative genomic studies. Methods Enzymol. 1993;224:294–309.
- Atienzar FA, Conradi M, Evenden AJ, et al. Qualitative assessment of genotoxicity using random amplified polymorphic DNA: comparison of genomic template stability with key fitness parameters in Daphnia magna exposed to benzo[a]pyrene. Environ Toxicol Chem. 1999;18:2275–2282.
- Chen Y, Schneeberger RG, Cullis CA. A site-specific insertion sequence in flax genotrophs induced by environment. New Phytol. 2005;167:171–180.
- Lolle SJ, Victor JL, Young JM, et al. Genome-wide non-Mendelian inheritance of extra-genomic information in Arabidopsis. Nature. 2005;434:505–509.