ABSTRACT
The technique of loop-mediated isothermal amplification (LAMP) utilizes 4 (5 or 6) primers targeting 6 (7 or 8) regions within a fairly small genome segment for amplification. This technique has a potential for greater specificity than two-primer methods, such as polymerase chain reaction. There are still no reports for primer–template mismatch of LAMP. In this study, a set of LAMP primers was designed, targeting the 16S–23S rRNA intergenic spacer region of Streptococcus dysgalactiae. The selectivity of the LAMP method was tested with 25 bacterial strains. There was a non-specific amplification when the genomic DNA of Streptococcus agalactiae was used as a template and it was indicated by a nucleotide basic local alignment search tool (BLAST) in GenBank. There were three false priming sites on backward inner primer and the internal primer–template mismatches extended the detection time from 21 min to 47 min. This study would be of great reference value for targeting sequence selection, primer design of LAMP and detection of antibiotic-resistant bacteria with LAMP.
Introduction
DNA polymerases catalyze the addition of nucleotides to the primer's 3′-OH end, as specified by complementarity to the template DNA.[Citation1] Mismatches between primers and targeted DNA can affect the duplex stability, which might hamper the ability of a system to amplify the template DNA.[Citation1] The amplification during polymerase chain reaction (PCR) can be prevented by a single mismatched site between the template and primer at the 3'-end of the primer, [Citation2–4] whereas the internal mismatches can be tolerated.[Citation5] The effects of the internal mismatches on the PCR efficiency have been studied and quantified.[Citation1,Citation6]
The technique of loop-mediated isothermal amplification (LAMP), developed and reported by Notomi et al. [Citation7] in 2000, utilizes 4 (5 or 6) primers targeting 6 (7 or 8) regions for amplification.[Citation8] This technique has a potential for greater specificity than two-primer methods, such as PCR. Based on the cost-effectiveness and specificity of LAMP, there has been a significant interest in its application towards basic researches in medical and environmental testing, point-of-care testing and diagnosis of infectious diseases.[Citation9–11] LAMP has been broadly applied in pathogen detection and has successfully detected Escherichia coli O157:H7, [Citation12] Actinobacillus actinomycetemcomitans, [Citation13] Mycobacterium tuberculosis, [Citation14] Streptococcus pneumoniae, [Citation15] Listeria monocytogenes, [Citation16] Staphylococcus aureus [Citation17,Citation18] and Streptococcus agalactiae.[Citation19] It has also been reported that a particular caution is exercised in LAMP to avoid mismatches at the 3′ ends of the primers. At those places, an elongation by the Bst DNA Polymerase is initiated. A limited number of mismatches (one or two, in the middle or at the 5′ ends of the primers) is thought to be acceptable, [Citation20] but to what extent such mismatches can affect the efficiency and specificity of the LAMP is still unknown.
The objective of this study was to investigate the effect of internal primer–template mismatches on the efficiency and specificity of LAMP. For this purpose, the 16S–23S rRNA intergenic spacer region of S. dysgalactiae, which had high similarities with that of S. agalactiae, was used as the targeting sequence.
Materials and methods
Primer design
A set of LAMP primers was designed and selected with PrimerExplorer 4 and Oligo 7, according to the reported methodology, targeting the 16S–23S rRNA intergenic spacer (GenBank Locus: EU860340.1) of S. dysgalactiae subsp. dysgalactiae strain NCTC 10238.[Citation21] The primers are listed in
Table 1. LAMP primers targeting the 16S–23S rRNA intergenic spacer of S. dysgalactiae.
Bacterial strains and DNA extraction
Twenty-five strains were used in this study (). Listeria strains were cultured overnight at 37 °C in DifcoTM Buffered Listeria Enrichment Broth Base (Becton, Dickinson and Company) and the other strains were cultured in Luria-Bertani broth (Sigma, St. Louis, MO, USA). DNA from the pure cultures was extracted with DNeasy® Blood & Tissue Kit (QIAGEN N.V.), according to the manufacturer's instructions. The DNA templates were used for determining the specificity of the designed LAMP primers. The amount of DNA template used was 100 pg per reaction. In the used negative control, the DNA template was substituted with Tris–ethylenediaminetetraacetic acid (EDTA) (TE) buffer.
Table 2. Specificity of the developed LAMP assay with primers targeting the 16S–23S rRNA intergenic spacer of S. dysgalactiae.
Specificity determination of LAMP assay
LAMP was performed in a 10 μL reaction mixture, which contained 0.8 mmol/L of each forward inner primer (FIP) and backward inner primer (BIP), 0.2 mmol/L of each F3 and B3 primers, 0.4 mmol/L of LF primer, 1.0 mmol/L dNTPs, 20 mmol/L Tris-HCl (pH 8.8), 10 mmol/L KCl, 10 mmol/L (NH4)2SO4, 6 mmol/L MgSO4, 0.1% (v/v) Triton X-100, 7.5% dimethyl sulphoxide (DMSO, v/v), [Citation11,Citation22] 1 × EvaGreen, 1 × Rox, 100 pg DNA template and 3.2 U Bst 2.0 WarmStart DNA Polymerase (New England Biolabs, Beverly, MA, USA).[Citation23] The reaction mixture was heated at 57 °C for 50 min in a StepOne™ System, 30 s per cycle, and the experiment was repeated three times. The equation ΔRn = Rn − Baseline was used, where Rn is the normalized reporter and ΔRn is the derivative reporter (relative fluorescence).
Determining the effect of primer–template mismatches on the efficiency of LAMP
Upon the results of specificity determination, the targeted sequence of S. dysgalactiae was aligned in GenBank with that of strains, detected as positive. This was done in order to locate the false priming sites and to determine the effect of the internal primer–template mismatches on the efficiency of LAMP.
PCR amplification and sequencing of mismatched template
In order to further verify the false priming sites, the forward outer primer F3 and the backward outer primer B3 of LAMP were used as upstream primer and downstream primer for PCR, respectively. The PCR was performed in a 20 μL reaction mixture with TaqMan® Universal Master Mix II (Life Technologies Corporation), according to the manufacturer's instructions: 10 μL TaqMan® Universal Master Mix II, 0.5 mmol/L of F3 and B3 primers and 100 pg DNA template of S. agalactiae ATCC 27956. The reaction mixture was held at 50 °C for 2 min and 95 °C for 10 min, followed by 40 cycles of denaturation for 15 s at 95 °C and 1 min anneal/extend at 60 °C. The amplified products were sequenced by Sangon Biotech (Shanghai) Co. Ltd., and were then compared with the sequence of S. agalactiae strain GBS6 in GenBank.
Results and discussion
Assaying selectivity
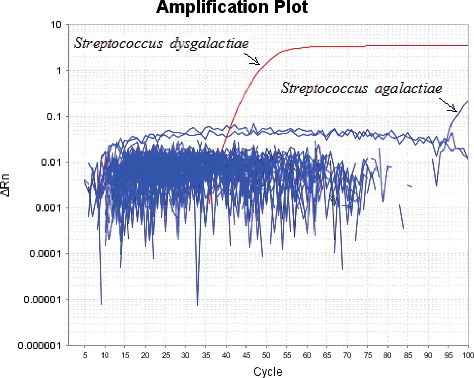
The LAMP assay with the designed primers was tested with 25 strains and, as shown in , only S. dysgalactiae ATCC 9542 and S. agalactiae ATCC 27956 were successfully detected. The results of the three repeated experiments were consistent, and there was no detectable false-positive response of the negative controls. The amount of DNA template added into each reaction was 100 pg, but the difference in the detection time between S. dysgalactiae ATCC 9542 and S. agalactiae ATCC 27956 was significant, as shown in . This result needed further analysis in terms of primers and target sequences.
Effect of primer–template mismatch on the efficiency of LAMP
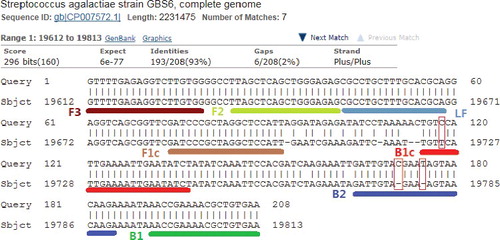
The used LAMP primers were targeted at a 208-bp sequence of S. dysgalactiae, as shown in . The identity between the targeted sequence of S. dysgalactiae and the partial sequence of most S. agalactiae strains was 193/208 (93%). In total, there were three false priming sites -- one site on primer B1c and two sites on B2. BIP was composed of B1c and B2 (B1c-TTTT-B2); therefore, the three false priming sites were only on primer BIP.
The three internal primer–template mismatches extended the detection time from 21 min to 47 min when the amount of DNA template was 100 pg per reaction.
Sequence of mismatched template
The sequencing results indicated that the amplified product's length of S. agalactiae ATCC 27956 was 202 bp. The sequences were 100% in concordance with the partial sequence of S. agalactiae strain GBS6. This verified that there were three false priming sites between LAMP primers and DNA template of S. agalactiae ATCC 27956, one site on primer B1c and two sites on B2, as indicated in
Final remarks
The effect of either 3′-terminal mismatches or internal mismatches on PCR has been extensively reported.[Citation1–4,Citation6] In this study, we reported the effect of three internal primer–template mismatches on LAMP for the first time. The results of the present study supported the hypothesis of Peyrefitte et al. [Citation20] that when there are three internal primer–template mismatches on the primer BIP, 100 pg DNA template of S. agalactiae ATCC 27956 can be amplified and detected within 50 min.
This study would be of great reference value for target sequence selection, primer design of LAMP, amplification time determination and detection of antibiotic-resistant bacteria with LAMP. The target sequence should be highly specific to avoid primer–template mismatch; if a limited number of false priming sites (less than three) cannot be avoided, the amplification should be controlled within 40 min to enhance the specificity; the mutant sites should not be designed in the middle or at the 5′ ends of the primers for detection of antibiotic-resistant bacteria with LAMP.
The LAMP assay also has a great potential for rapid detection of resistant mutants after an antibiotic administration, when the mutant sites are designed at the 3′ ends of the primers. The main advantage of the PCR method over routine culture methods is the shorter turnaround time for detecting methicillin-resistant S. aureus, [Citation24] whereas, compared to real-time PCR, the LAMP method is more cost-effective and provides an excellent availability for rapid examination in a hospital clinical laboratory.[Citation23]
Conclusions
The present study indicated that three internal primer–template mismatches on BIP of LAMP extended the detection time from 21 min to 47 min, which verified the effect of primer–template mismatches on the efficiency of LAMP for the first time. This study may be of great value for target sequence selection, primer design of LAMP, amplification time determination and detection of antibiotic-resistant bacteria with LAMP. The LAMP assay may also be used for rapid detection of resistant mutants after an antibiotic administration, when the mutant sites are designed at the 3' ends of the primers. Also, the LAMP method is cost-effective and provides an excellent availability for rapid examination in a hospital clinical laboratory.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bru D, Martin-Laurent F, Philippot L. Quantification of the detrimental effect of a single primer-template mismatch by real-time PCR using the 16S rRNA gene as an example. Appl Environ Microbiol. 2008;74:1660–1663.
- Ayyadevara S, Thaden JJ, Shmookler Reis RJ. Discrimination of primer 3'-nucleotide mismatch by taq DNA polymerase during polymerase chain reaction. Anal Biochem. 2000;284:11–18.
- Day JP, Barany F, Bergstrom D, et al. Nucleotide analogs facilitate base conversion with 3' mismatch primers. Nucleic Acids Res. 1999;27(8):1810–1818.
- Huang MM, Arnheim N, Goodman MF. Extension of base mispairs by Taq DNA polymerase: implications for single nucleotide discrimination in PCR. Nucleic Acids Res. 1992;20(17):4567–4573.
- Kwok S, Kellogg DE, McKinney N, et al. Effects of primer-template mismatches on the polymerase chain reaction: human immunodeficiency virus type 1 model studies. Nucleic Acids Res. 1990;18(4):999–1005.
- Christopherson C, Sninsky J, Kwok S. The effects of internal primer–template mismatches on RT-PCR: HIV-1 model studies. Nucleic Acids Res. 1997;25(3):654–658.
- Notomi T, Okayama H, Masubuchi H, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63.
- Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes. 2002;16:223–229.
- Fakruddin M. Loop mediated isothermal amplification (LAMP) – an alternative to polymerase chain reaction (PCR). Bangladesh Res Publ J. 2011;5:425–539.
- Khiyami MA, Almoammar H, Awad YM, et al. Plant pathogen nanodiagnostic techniques: forthcoming changes? Biotechnol Biotechnol Equip. 2014;28(5):775–785.
- Wang Deguo. Novel primers for increased specificity and sensitivity for the detection of Staphylococcus aureus by real-time LAMP. CyTA. J Food. 2015;14(1):88–91.
- Maruyama F, Kenzaka T, Yamaguchi N, et al. Detection of bacteria carrying the stx2 gene by in situ loop-mediated isothermal amplification. Appl Environ Microbiol. 2003;69:5023–5028.
- Osawa R, Yoshida A, Masakiyo Y, et al. Rapid detection of Actinobacillus actinomycetemcomitans using a loop-mediated isothermal amplification method. Oral Microbiol Immunol. 2007;22:252–259.
- Iwamoto T, Sonobe T, Hayashi K. Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. J Clin Microbiol. 2003;41:2616–2622.
- Seki M, Yamashita Y, Torigoe H, et al. Loop-mediated isothermal amplification method targeting the lytA gene for detection of Streptococcus pneumoniae. J Clin Microbiol. 2005;43:1581–1586.
- Tang MJ, Zhou S, Zhang XY, et al. Rapid and sensitive detection of Listeria monocytogenes by loop-mediated isothermal amplification. Curr Microbiol. 2011;63:511–516.
- Hwang SY, Kim SH, Jang EJ, et al. Novel multiplex PCR for the detection of the Staphylococcus aureus superantigen and its application to raw meat isolates in Korea. Int J Food Microbiol. 2007;117:99–105.
- Lim KT, Ju Teh CS, Thong KL. Loop-mediated isothermal amplification assay for the rapid detection of Staphylococcus aureus. Biomed Res Int. 2013;2013:895816.
- Kimura K, Yanagisawa H, Wachino JI, et al. Rapid and reliable loop-mediated isothermal amplification method for detecting Streptococcus agalactiae. Jpn J Infect Dis. 2013;66(6):546–548.
- Peyrefitte CN, Boubis L, Coudrier D, et al. Real-time reverse-transcription loop-mediated isothermal amplification for rapid detection of rift valley fever virus. J Clin Microbiol. 2008;46:3653–3659.
- Li SL, Zhang XB, Wang DG, et al. Simple and rapid method for detecting foodborne Shigella by a loop-mediated isothermal amplification. J Rapid Methods Autom Microbiol. 2009;17(4):465–475.
- Frackman S, Kobs G, Simpson D, et al. Betaine and DMSO: enhancing agents for PCR. Promega Notes. 1998;65:27.
- Misawa Y, Yoshida A, Saito R, et al. Application of loop-mediated isothermal amplification technique to rapid and direct detection of methicillin-resistant Staphylococcus aureus (MRSA) in blood cultures. J Infect Chemother. 2007;13:134–140.
- Baddour MM, AbuEIKheir MM, Fatani AJ. Comparison of mecA polymerase chain reaction with phenotypic methods for the detection of methicillin-resistant Staphylococcus aureus. Curr Microbiol. 2007;55:473–479.