ABSTRACT
Panax stipuleanatus Tsai is a type of medicinal plant within north-west Vietnam. In this study, inter simple sequence repeat markers were employed to investigate the genetic diversity of naturally distributed populations classified by habitat for this species. Genetic diversity at the species level was moderate (HeT = 0.254; PPBT = 96.02%). Genetic diversity was not equal in two populations. The result showed higher population genetic diversity in the Lao Cai region (HeBX = 0.266; PPBBX = 91.48%) as compared to those located in Lai Chau region (HeHT = 0.235; PPBHT = 84.66%). The interpopulation gene differentiation was small (GSTP = 0.03) with the genetic distance among populations was DP = 0.0103. Gene flow within populations was as high as Nm = 7.36.
Introduction
Panax stipuleanatus Tsai is a type of medicinal plant which is naturally distributed within north-west Vietnam. The Panax species populations grow in small groups scattered amongst the herbaceous storey of primary, closed, evergreen, seasonal, tropical, broad-leaved forests on sandy and shale soils. The soil structure is wet and well-drained, and the area is located at an altitude between 1600 and 2400 m over the two regional areas: Ho Thau (HT; Lai Chau Province) and Bat Xat (BX; Lao Cai Province). The two populations are very well adapted to the tropical monsoon climate associated with these particular mountainous localities.[Citation1,Citation2] In the two investigated populations, mature individuals have been harvested and used by the native population as treatment for several acute medical issues and general human health maintenance for centuries. Due to the over-exploitation of these medicinal plant's rhizomes for medical use and the eradication of their primary growing habitat through forest clearance, which is to be used for Amomum tsao-ko plantation, has led to the loss of genetic biodiversity and as a result the studied plant was classified as critically endangered under the national category with criteria of A1c,d.[Citation2,Citation3]
Currently, this species is considered to be endangered and its distribution is limited. There are two indigenous populations for this species that have been identified within north-west Vietnam. During the year 2013 and 2014, field identification research has identified approximately 100–150 individual plants remaining in the two regions studied. However, studies on the genetic diversity of this species have not been conducted in Vietnam. Reduction in genetic diversity is an actual risk to P. stipuleanatus. Genetic variation is currently understood as a critical variable to the long-term survival of a population or species.[Citation4,Citation5] Understanding the genetic variation and diversity within and among populations of rare and endangered species is essential when developing management strategies for both in situ and ex situ conservation activities.[Citation6] Thus, estimating inter- and intra-population genetic diversity is critical to the protection and long-term availability of P. stipuleanatus both in terms of ecological biodiversity and medically related uses. Current research methods support the use of molecular markers as suitable and accurate tools for population genetic diversity detection.
The advantages of inter simple sequence repeat (ISSR) lies within in its low-cost use, convenience of use and high level of reliability in reproducing results.[Citation7–10] As such, ISSR is used widely and is accepted as a tool in population genetic studies of both wild and cultivated plants.[Citation9]
In this study, the ISSR marker system was employed to induce DNA fingerprints for the estimation of genetic diversity and the identification of genetic differentiation in wild P. stipuleanatus populations found and distributed in their natural habitats. The objectives of this study were as follows: (1) to estimate genetic diversity; (2) to analyse genetic relationships and differentiation among two populations and (3) to contribute and catalogue the data of this study for use in the conservation and sustainable utilization of the researched medicinal plants within Vietnam.
Materials and methods
Plant materials
From December 2013 to May 2014, a total of 60 individuals presenting natural populations of the species from Ban Giang commune, Ho Thau District (Lai Chau Province) and Muong Hum commune, Bat Xat District (Lao Cai Province), which corresponded to two naturally distributive populations of this species, were sampled across their original habitats (). Chosen individuals for sampling were separated from each other at a distance of at least 50 m.
Table 1. Geographic localities of P. stipuleanatus populations in this study.
Fresh leaves were collected, dried in sealed bags with silica gel and brought to the laboratory, where each sample was preserved at a constant –20 °C for DNA analysis.
DNA extraction and ISSR-PCR amplification
Total genomic DNA was extracted using cetyltrimethyl ammonium bromide (CTAB) protocol I [Citation11] with a modification of adding 10% SDS to the extraction buffer, which was then dissolved in water for the subsequent use. ISSR primers used in this study were synthesized by Bioneer Corporation (Republic of Korea), according to the primer set published by the University of British Columbia and Zagazig University (Egypt). Sixty ISSR primers were initially screened, and 17 of them, which yielded bright, clear bands and at least possessed one polymorphic band in both populations, were used for the analysis of all 60 samples (). PCR amplification was repeated for those working primers to check the stability and reproducibility of ISSR DNA fingerprinting.
Table 2. ISSR primers used in this study.
PCRs were performed in 20 µl reactions containing 2 mmol/L MgCl2, 0.25 mmol/L each of dNTPs, 1U Taq DNA polymerase (ThermoScientific), 0.2 µmol/L primer and approximately 30 ng DNA templates. The amplifications were performed in a Peqstar 96X Universal Gradient thermocycler (PEQLAB Biotechnologie GmbH, Germany) with the following program: initial denaturation at 94 °C for 5 min; 10 cycles of 94 °C for 45 s, annealing temperature +5 (Ta +5) °C () for 45 s, decreased 0.5 °C/cycle, 72 °C for 1 min 30 s; 36 cycles of 94 °C for 45 s, annealing temperature for 45 s, 72 °C for 1 min 30 s; final extension at 72 °C for 15 min; the amplification products were separated in 2% agarose gel, using TBE buffer at 60 V for 3 hours, stained with ethidium bromide (0.5 µg/ml), and photographed under 254/312 nm wavelength lights using Micro Doc Gel Documentation System (Cleaver Scientific, USA).
Data analysis
Since ISSR markers are dominantly inherited, each band was assumed to represent the phenotype at a single biallelic locus.[Citation12] ISSR bands were scored as presence (1) or absence (0) characters, to construct the binary data matrix. Microsoft Office Excel 2007 was used to estimate genetic diversity parameters: the percentage of polymorphic bands (PPB).[Citation13] The average expected heterozygosity (He = ΣjLhj/L) was tested using Nei's gene diversity statistics.[Citation13] The gene differentiation among populations was calculated as GST = (HeT – HST)/HeT. Gene flow was estimated using the formula: Nm = (1–GST)/4 GST.[Citation11] The Nei's genetic distance between populations was calculated as: DXY = –ln(IXY).[Citation14] Similarity coefficient between pair of samples and unweighted pair group method with arithmetic mean (UPGMA) dendrogram for genetic relationship among studied samples was calculated and established by using NTSYSpc 2.1 (Numerical Taxonomy and Multivariate Analysis System) software.[Citation15]
Results and discussion
Genetic diversity
The 17 selected primers yielded 176 reproducible bands for total investigated samples, 166 reproducible bands with BX population, 170 reproducible bands with HT population. For species level, the number of bands per primer varied between 7 (UBC 856T) and 14 (17899B), with an average of 10.4. In BX population, the number of bands per primer varied between 6 (UBC 807) and 14 (17899B), with an average of 9.8. In HT population, the number of bands per primer varied between 7 (UBC 856T) and 14 (17899B), with an average of 10 ().
Genetic diversity at the species level were moderate, the estimated heterozygosity and percentage of polymorphic bands were HeT = 0.254; PPBT = 96.02%, respectively. Among the two investigated populations, HT population possessed the lower level of genetic diversity (HeHT = 0.235; PPBHT = 84.66%), while BX population harboured higher level (HeBX = 0.266; PPBBX = 91.48%).
Genetic relationship
Among the two investigated populations, the interpopulation gene differentiation was significantly small (GSTP = 0.03) with the genetic distance among populations was DP = 0.0103, which means a 3% differentiation among populations exist. The gene flow within populations was as high as Nm = 7.36 showed that the migration among the two populations was high.
The gene similarity coefficients among the species were varied, ranging from 0.483 to 0.943 with a mean of 0.728 (). In HT population, they ranged from 0.534 to 0.943 with a mean of 0.746; and in BX population they ranged from 0.483 to 0.926 with a mean of 0.708.
The ISSR markers in this study yielded reproducible polymorphic bands in 60 individuals, which were used to investigate samples belonging to two populations of P. stipuleanatus in the north-west Vietnam, which was classified by habitat. This method provides a highly effective and reliable molecular-level tool for analysation of genetic diversity and genetic relationships within the species. This study reports the genetic diversity at species and population levels. The extent of genetic variation within two natural populations, the gene differentiation and the genetic distance among them were showed.
Wide ranges of species possess the life history traits of dicotyledon (long-lived perennial life form, narrow geographic range, outcrossing breeding system and ingested seed dispersal mechanism), which are similar to P. stipuleanatus (found in the north-west Vietnam) were researched. Hamrick and Godt,[Citation16] reported that the genetic diversity based on allozyme were PPB = 42%--46%, He = 0.10–0.14, GST = 0.14--0.24. And Nymbom,[Citation17] based on RAPD, reported that the genetic diversity were He = 0.19–0.24, GST = 0.17–0.23. Thus, the results from the current research showed that P. stipuleanatus in the north-west Vietnam possessed higher genetic diversity but the interpopulation gene differentiation was significantly small. Achieved results showed higher population genetic diversity related to PPB and heterozygosity compared to previous studies based on RAPD,[Citation18–20] on Allozyme, [Citation6,Citation21] on AFLP,[Citation14,Citation22] on DALP [Citation23] in other Panax populations. However, the similarity coefficients among the pair of samples in this study were higher,[Citation24] and the expected heterozygosity of investigated populations were lower in the case of natural Panax ginseng using SSR markers,[Citation25] which showed the limitations of ISSR markers in individual discrimination.
Using the same technique as in this study to induce DNA fingerprinting in P. ginseng cultivated in north-east China, Ref. [Citation26] reported that the genetic diversity was high at the species level (He = 0.2886; PPB = 98.96%) but relatively lower at the cultivated-type level: in garden ginseng the reported results were He = 0.229, PPB = 85.42%; in forest ginseng the reported results were He = 0.170, PPB = 57,29%; and in transplanted wild ginseng the reported results were He = 0.202, PPB = 76.04%. Genetic differentiation was also detected among populations of garden ginseng at GST = 0.319 with Nm = 1.069; forest ginseng was GST = 0.233 with Nm = 1.648; and transplanted wild ginseng was GST = 0.2540 with Nm = 1.467. Compared to these results, this study revealed that genetic diversity of P. stipuleanatus in north-west Vietnam contained lower levels of genetic diversity as compared to P. ginseng in north-east China. The genetic differentiation among populations of investigated species was lower than among populations belonging to three types of ginseng as previously investigated in the study from [Citation26]. The gene flows amongst the populations from this study were significantly higher than those amongst the populations investigated; the high level of genetic diversity in this study can be attributed to the investigated species evolutionary development. Due to the long lifespan and overlapping generations of the populations within the prior study, considerable genetic variability has been accumulated and conserved under various selection traits during the evolutionary process.[Citation26] Beyond the studies, the local people have been using traditional methods to select mature ginseng for harvest and save young ginseng from the natural protected areas. This is possibly due to the fact that the harvested and conserved ginseng can be reproduced by seed, which facilitates to a certain degree the genetic diversity of the local ginseng populations.
The genetic diversity of the two studied wild populations is unequal. BX population possessed the higher genetic diversity as compared to this in HT population, which reflects the fact that recently P. vietnamensis var. fuscidiscus, which is more economically valuable in Vietnamese and Greater Asian medicinal plant markets, was discovered in Lai Chau province and people have overharvested all taxa of Panax here for economic purpose, therefore there has been use of both P. stipuleanatus rhizomes as adulterants of P. vietnamensis and its variety.
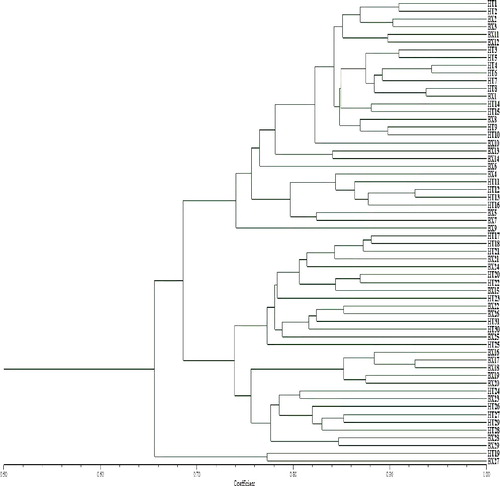
The UPGMA dendrogram for genetic relationship of total investigated samples and for each of two investigated populations is shown in . Using 17 ISSR primers, the samples were clustered by regional population. This was confirmed by the results on the interpopulation gene differentiation and the genetic distance among populations. The level of gene differentiation among populations can be closely related to various factors such as the long-term evolutionary history and the nature of species, difference in breeding systems, gene flow in conditions of habitat fragmentation, population isolation etc.[Citation6,Citation16,Citation27,Citation28] Thus, the low genetic differentiation among populations of the P. stipuleanatus in this study may be the consequence of the strong genetic crossing nature of the species, the abundance of pollinators (insects) and dispersive animals (rodents and birds), insufficient partitioned terrain such that gene flows occur, and other biological traits.
Conclusions
The understanding on population genetic variability is essential to effective conservation and sustainable management. The medium genetic diversity at population level, relatively high at species level and the high gene flow are the advantages for conservation and development of the ginseng species in North Vietnam. However, the uncontrolled overharvesting of these species without an actionable conservation strategy may lead to the increased reduction of genetic diversity and reserves of this species, much more so than indicated by the disproportion of population genetic diversities between the two habitats. Thus, it is of critical importance to further investigate those species for conservation purposes and for sustainable harvesting and use as valuable natural resources.
Acknowledgments
We would like to thank Mr Le Van Gioi for his help with the field investigation and materials collection.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Tap N. The species of Panax L. in Vietnam. J Med Mater. 2005;10:71–76. Vietnamese.
- Tap N. Handbook of medicinal plants protect. Vietnam Non-Timber Forest Products Network; 2007. Vietnamese.
- Vietnam Academy of Science and Technology. Vietnam red data book, Part II. Plants. Hanoi: Natural Science and Technology Publish House; 2007.
- Anatonovis J. Genetic variation within population. In: Dirzor R, Sarukan J, editors. Perspectives on plant population biology. Sunderland: Sinauer; 1984. p. 229–241.
- Beardmore JA. Extinction, survival and genetic variation. In: Schoenwald-Cox CM, Chamber SM, Macbryde B, Thomas L, editors. Genetics and conservation. Menlo Park (CA): Benjamin-Cummings; 1983. p. 125–151.
- Hogbin PM, Peakall R. Evalution of the contribution of the genetic research to the management of the endangered plant Zieria prostrate. Conserv Biol. 1999;13:514–522.3
- Lu X, Liu L, Gong Y, et al. Cultivar identification and genetic diversity analysis of broccoli and its related species with RAPD and ISSR markers. Sci Hortic. 2009;122:645–648.
- Nagoaka T, Ogihara Y. Applicability of inter-simple sequence repeat polymorphism in wheat for use as DAN markers in comparison to RFLP and RAPD markers. Theor Appl Genet. 1997;93:133–139.
- Roy SC, Chakraborty BN. Genetic diversity and relationships among tea (Camellia sinensis) cultivars asrevealed by RAPD and ISSR based fingerprinting. Indian J Biotechnol. 2009;8:370–376.
- Zietkiewicz E, Rafalski A, Labuda D. Genome fingerprinting by simple sequence repeat (SSR) -anchored polymerase chain reaction amplification. Genomics. 1994;20:176–183.
- Weising K, Nybom H, Wolff K, et al. DNA fingerprinting in plants principles, methods, and applications. 2nd ed. London: CRC Press; 2005.
- Williams JGK, Kubelik AR, Livak KJ, et al. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:31–65.
- Nei M. Estimation of average teterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590.
- Vicente MCD, Lopez C, Fulton T. Genetic diversity analysis with molecular marker data: learning module. Rome: International Plant Genetic Resources Institute (IPGRI) and Cornell University; 2003.
- Rohlf FJ. NTSYSpc-Numerical taxonomy and multivariate analysis system version 2.1-user guide. New York: Applied Biostatistics Inc.; 2004.
- Hamrick JL, Godt MJW. Effects of life history traits on genetic diversity in plant species. Phil Trans R Soc B. 1996;35:1291–1298.
- Nybom H, Bartish IV. Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Perspect Plant Ecol Evol Syst. 2000;3:93–114.
- Artyukova EV, Kozyrenko MM, Koren OG, et al. RAPD and allozyme analysis of genetic diversity in Panax ginseng C.A. Meyer and P. quinquefolius L. Russian J Genet. 2004;40:178–185.
- Obae SG, West TP. Effects of anthropogenic activities on genetic diversity and population structure of American ginseng (Panax quiuefolius L.) growing in West Virginia. J Hortic For. 2011;3(9):270–281.
- Schlag EM, McIntosh MS. RAPD-based assessment of genetic relationships among and within American ginseng (Panax quinquefolius L.) populations and their implications for a future conservation strategy. Genet Resour Crop Evol. 2012;59:1553–1568.
- Jennifer MC, Hamrick JL. Genetic diversity in harvested and protected populations of wild American ginseng, Panax quinquefolius L. (Araliaceae). Am J Bot. 2004;91(4):540–548.
- Zhuravlev YN, Reunova GD, Kats IL, et al. Research genetic variability and population structure of endangered Panax ginseng in the Russian primorye. Chin Med. 2010;5:21.
- Cui XM, Xiao H, Yang JJ, et al. DALP analysis on genetic diversity of Panax notoginseng. Chin Med. 2014;5:123–129.
- Bai D, Brandle J, Reeleder R. Genetic diversity in North American ginseng (Panax quinquefolius L.) grown in Ontario detected by RAPD analysis. Genome. 1997;40:111–115.
- Reunova GD, Koren OG, Muzarok TI, et al. 2014. Microsatellite analysis of Panax ginseng. Natural populations in Russia. Chin Med. 2014;5:231–243.
- Li S, Li J, Yang XL, et al. Genetic diversity and differentiation of cultivated ginseng (Panax ginseng C. A. Meyer) populations in north-east China revealed by inter-simple sequence repeat (ISSR) markers. Genet Resour Crop Evol. 2011;58:815–824.
- Hamrick JL, Godt MJW. Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS, editors. Plant population genetics, breeding and germplasm resources. Sunderland: Sinauer Associates; 1989. p. 43–63.
- Schaal BA, Hayworth DA, Olsen KM, et al. Phylogeographic studies in plants: problems and prospects. Mol Ecol. 1998;7:465–474.