ABSTRACT
Economically important representatives of the Artemisia genus have wide application in pharmaceutics, landscape architecture and agriculture. The aim of this study was to phylogenetically analyse the 18S-26S rDNA, the internal transcribed spacer (ITS) nucleotide sequences of 19 Artemisia samples collected from Ordu Province of Turkey. Analyses were conducted using neighbour-joining (NJ), maximum-parsimony (MP) and maximum-likelihood (ML) algorithms. Our analysis revealed two unique haplotypes within our samples, including a rare one (Haplotype-I, represented by a single sample) and a common one (Haplotype-II, represented by 18 samples). In all trees, both of our haplotypes appeared in the same lineage with Artemisia sylvatica, Artemisia argyi and Artemisia verlotiorum. Although rDNA-ITS is known to be a variative marker, oddly it was highly conserved in Artemisia, which is why we used this marker to determine the phylogenetic affiliation of the analysed plants.
KEYWORDS:
Introduction
Artemisia is the largest genus in the Anthemideae tribe of Asteraceae family and covers more than 500 taxa which are at species or subspecies levels. In spite of the harmful effects of some of its species (invading nurseries and farmlands; toxic and allergenic for humans), the Artemisia genus has a wide area of usage in different fields, including pharmaceutics, landscape architecture and agriculture.[Citation1,Citation2] For instance, Artemisia vulgaris was used for flavouring beer before hobs [Citation1], additionally its leaves have been used for human consumption [Citation3] and its extracts have been used as a cure for some gynaecological problems and as mosquito repellents.[Citation4,Citation5] Besides A. vulgaris, other Artemisia species have economic importance, too. For example, Artemisia annua and Azadirachta indica have been used as antimalarial agents in China and Thailand for a long time.[Citation6,Citation7] Despite its economic and ecological importance, there are still controversies in the taxonomy of Artemisia. Conventionally, the Artemisia L. genus is divided into five subgenera: Artemisia, Absinthium (Mill.) Less., Dracunculus (Besser) Rydb, Seriphidium Besser ex Less. and Tridentatae (Rydb.) McArthur, based on some morphological characters, including the type of capitula and floret fertility. Recently, a sixth subgenus, Pacifica, from Hawaii has been suggested.[Citation8–10] On the other hand, molecular phylogenetic studies revealed that this classification does not entirely reflect the natural evolutionary relationships within the genus. [Citation11] But because of the lack of polyphasic studies concerning both morphological and molecular data, this classification system is still valid.[Citation11,Citation12] The genus is widely distributed across the Northern Hemisphere but is rare (about 10 species) in the Southern Hemisphere; and Eurasia is the centre of origin.[Citation8,Citation13]
Several molecular methods have been used to determine the genetic diversity and relationships among different Artemisia species, including karyotyping,[Citation14] cpDNA restriction site variation analysis,[Citation15] polymerase chain reaction–restriction fragment length polymorphism (PCR--RFLP) analysis of several genes,[Citation16,Citation17] microsatellite polymorphism analysis [Citation18,Citation19] and Random amplified polymorphic DNA (RAPD) analysis.[Citation20–22]
To the best of our knowledge, none of the Artemisia species in Turkey has been verified with molecular phylogenetic studies providing nucleotide sequence data, even though 26 taxa (including 21 species) within four subgenera (Artemisia, Absinthium, Dracunculus and Seriphidium) have been reported there, so far.[Citation23] Thus, the aim of this study was to assess the genetic diversity of Artemisia species in Ordu Province of Turkey in an attempt to validate the formerly reported species from this area, using molecular techniques. Additionally, the first molecular data from Turkey will be submitted to the international databases.
Materials and methods
Plant materials
Nineteen samples of Artemisia species were collected from different geographic locations in Ordu Province of Turkey (). After fresh Artemisia plants were collected from each sampling site, they were stored in plastic containers in a refrigerator until DNA extraction.
Table 1. Geographic origins of the Artemisia samples collected in this study and EMBL accession numbers for the ITS nucleotide sequences of the respective haplotypes.
DNA extraction and PCR
Genomic DNA was extracted from fresh Artemisia samples using the method of Haymes [Citation24] with some modifications. Briefly, 20 mg of leaf tissue were quickly crushed using a hand pestle by adding liquid nitrogen into the eppendorf tube. Then, 500 µL of extraction buffer was added to the grinded leaf tissue. The mixture was incubated in a shaking dry block heater for 30 min at 65 °C and 200 µL of chloroform/isoamylalcohol (24/1) was transferred to the mixture. After centrifugation (Kuboto 3500) for 5 min at 10,000 r/min, the supernatant was transferred to a new eppendorf tube and 600 µL of ethanol/acetate (96/4) was added for precipitating the DNA; then the DNA was washed with 300 µL of ethanol (70%). Next, 100 µL of nuclease-free water was added to the white pellet; 1 µL of RNAse (Qiagen)100 mg/mL) was added to resuspended DNA and the samples were incubated for 1 h at 37 °C. DNA was storaged at −20 °C until later use.
PCR amplification was performed in a 25 μL volume containing: 1×PCR buffer, 10 mmol/L deoxyribonucleoside triphosphates (dNTPs), 2.5 mmol/L of MgCl2, 10 mmol/L of primers ITS1 (internal transcribed spacer) and ITS4, [Citation25] 40 ng of template DNA and 5 U Taq polymerase (Thermo Scientific, Maxima Hot Start). The PCR procedure was performed using a thermal cycler (PeqLab) as follows: initial denaturation at 95 °C for 5 min, followed by 35 cycles of 95 °C for 1 min, 48.7 °C for 2 min and 72 °C for 2 min, concluding with a final extension step at 72 °C for 7 min. The PCR products were electrophoresed in a 1.5% agarose gel prepared in 1×TBE (Tris-borate-ethylenediaminetetraacetic acid) buffer. After staining with ethidium bromide (1 mg/mL), gels were visualized using the QUANTUM ST5 (Vilber Lourmat) gel documentation system.
Nucleotide sequencing and phylogenetic analysis
The amplified DNA was sequenced commercially by Macrogen Inc. (Amsterdam, the Netherlands) from both strands, using the primer set ITS1/ITS4. Sequenced fragments were assembled using BioEdit.[Citation26] Multiple nucleotide alignments of our new rDNA-ITS haplotypes and the haplotypes of different Artemisia species (from GenBank) with the highest BLAST (Basic Local Alignment Search Tool in NCBI) scores were made using ClustalX [Citation27] and edited with BioEdit.[Citation26] The Akaike information criterion (AIC) [Citation28] and Bayesian information criterion (BIC) tests were performed using jModelTest v. 0.1 package program [Citation29,Citation30] to determine the best-fitting evolutionary model for our data-set. To reveal phylogenetic relations, neighbour-joining (NJ),[Citation31] maximum-parsimony (MP) [Citation32,Citation33] and maximum-likelihood (ML) algorithms were used. NJ and MP analyses were made using the software program PAUP* v.4.0b10, [Citation34] whereas PhyML 3.0 [Citation29] was used for ML. The heuristic search approach was applied for the MP analysis using the tree bisection and reconnection swapping algorithm with 10 random repetitions. To test the reliability of the trees, bootstrap tests [Citation35,Citation36] were conducted with 1.000 pseudo replications for MP and ML and 10.000 pseudo replications for the NJ analysis. The nucleotide sequence similarity and the evolutionary divergence among haplotypes were evaluated using BioEdit and MEGA version 6, [Citation37] respectively.
All haplotype sequences obtained in this study were deposited in the European Molecular Biology Laboratory (EMBL) data bank (https://www.ncbi.nlm.nih.gov/genbank/update/) under accession numbers KU523875–KU523893 ().
Results and discussion
Phylogenetic analysis based on the nucleotide sequences of transcript-coding or non-coding loci has become the major tool for determining the genetic diversity among organisms in the last two decades. On the other hand, revealing the phylogenetic associations of closely related species or populations can only be possible by using fast-evolving DNA regions as markers.[Citation38] From this point of view, nuclear rDNA internal transcribed spacer (rDNA-ITS) regions are useful molecular markers for reconstructing phylogenies at the inter- or intra-specific level because they evolve fast and at nearly neutral rates.[Citation39] Beginning with the pioneering study of Kornkven et al. [Citation40], this marker has been commonly used for Artemisia and, hence, rDNA-ITS haplotypes of different Artemisia species are available in international databases (e.g. GenBank). For this reason, in our study, we sequenced the full length of 18S-26S rDNA-ITS (approximately 700 bp) of the Artemisia samples collected from Ordu Province as shown in
We determined two haplotypes among our samples. Haploype-I was represented by one sample (F2), whereas the rest of our samples (G1, G3, G4, G6; F4, F5, F6; P4, P5, P6; O1, O3, O4, O6; U1, U2, U3, U4) shared Haplotpe-II. Although molecular evidence points out some conflicts, the Artemisia L. genus is traditionally divided into six subgenera namely, Artemisia, Absinthium (Mill.) Less., Dracunculus (Besser) Rydb., Seriphidium Besser ex Less., Tridentatae (Rydb.) McArthur and the recently identified Pacifica.[Citation8,Citation10,Citation12,Citation41] Because BLAST results suggested that our haplotypes are related to subgenus Artemisia, we established a data-set comprising closely related species of this subgenus. Phylogenetic analyses were performed over 604 alligned nucleotides with 35 segregating sites. Because both AIC and BIC tests suggested a Tamura--Nei plus Gamma (TrNef+G) substitution model (G: 0.016), NJ and ML trees were drawn with this model. Parsimony analysis was conducted using 21 synapomorphic characters and revealed two most parsimonious trees with 43 steps (consistency index CI: 0.884; retention index (RI): 0.919; homoplasies index (HI): 0.116). The phylogenetic trees drawn with NJ, MP and ML algorithms showed similar topologies with minor differences. In all of them, our haplotypes were placed in the same lineage with Artemisia sylvatica, Artemisia argyi and Artemisia verlotiorum (Figure 1). Haplotype-I appeared as sister to A. sylvatica with 98.6% nucleotide sequence similarity and the pairwise genetic divergence was 0.016. This relation was supported with a/the 50% bootstrap value in the NJ tree. On the other hand, Haplotpe-II appeared as sister to the lineage above and this node was also supported with a/the 50% bootstrap value in the NJ tree. The nucleotide sequence similarity and genetic divergence between Haplotype-II and Haplotype-I were 99.7% and 0.002, between Haplotype-II and A. sylvatica were 98.8% and 0.013. Oddly, two other species of this lineage, A. argyi and A. verlotiorum, showed the same rDNA-ITS haplotype and appeared as sister to the haplotypes above. This relation was supported with 96%, 85% and 95% bootstrap values in the NJ, MP and ML trees, respectively. Although, rDNA-ITS is expected to be a highly variable locus between different species or even between populations, this was not the case in the Artemisia genus because different Artemisia species showed the same haplotype (e.g. Artemisia integrifolia/Artemisia princeps and Artemisia montana) as mentioned above ( and ). Additionally, nucleotide sequence similarities between close species sharing the same lineage were mostly higher than 99.5% as shown in
Figure 1. NJ tree showing the phylogenetic relationships between 18S-26S rDNA-ITS haplotypes obtained in this study (Haplotype-I and -II) and haplotypes of closely related species of Artemisia subgenus (obtained from GenBank). Bootstrap values (higher than 50%) from NJ, MP and ML analysis are given in this order.
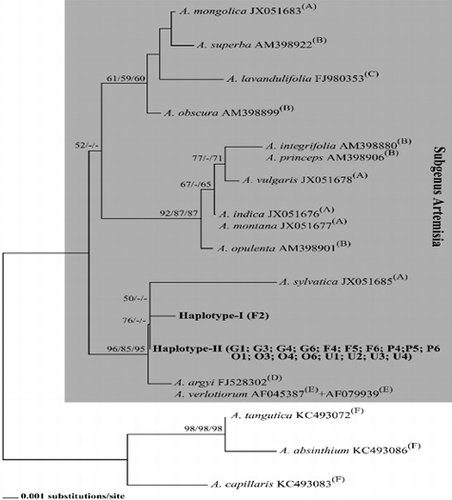
Table 2. Pairwise estimate of evolutionary divergence (shaded in grey) and nucleotide sequence percentage similarities between the ITS haplotypes obtained in this study and related Artemisia species from GenBank.
A lot of research has been carried out to better understand the phylogenetic analysis of the genus Artemisia and its relationships to the other (four) subgenera, Absinthium, Dracunculus (Besser) Rydb, Seriphidium Besser ex Less. and Tridentatae (Rydb), in different parts of the world. The phylogenetic relationship among the different Artemisia species collected from different regions of Pakistan based on the chloroplast gene RPS11 was reported by Mahmood et al. [Citation17]. Molecular phylogenetic analyses of Hawaiian Artemisia and its worldwide divergence based on nuclear and chloroplast DNA markers were reported by Hobbs and Baldwin.[Citation10] As discussed by Haghighi et al. [Citation42], the phylogenetic relationships among Artemisia species based on nuclear ITS and chloroplast psbA-trnH DNA markers using three sections of Artemisia, Dracunculus and Serphidium propose that the ITS and cppsbA-trnH markers are practicable in the systematic revision of troubled taxa at the intra-genus level in plants. Furthermore, Pellicer et al. [Citation11] performed phylogenetic analysis of the annual Artemisia within its major lineages and suggested that annual Artemisia have been specially misidentified at a subgeneric level and verified that they are phylogenetically restricted to basal grades.
Artemisia species reported in Turkey have been generally verified with microscopic analyses and cytogenetic studies reported by Kurşat et al. [Citation23,Citation43,Citation44] and Tabur et al. [Citation45]. However, to the best of our knowledge, none of these Artemisia species have been verified with molecular phylogenetic studies based on nucleotide sequence data in Turkey, so far. Here we used phylogenetic assays based on nucleotide sequence data to better understand the genetic diversity of Artemisia species collected from plant samples in Ordu Province of Turkey. Our data that Haplotype I was grouped separately from Haplotype II indicate that further polyphasic studies including a larger number of samples are needed to further explore whether these might represent different subspecies.
Conclusions
To the best of our knowledge, this is the first study to report molecular phylogenetic analyses of the genus Artemisia in Ordu Province of Turkey. In the light of the available molecular data, each of the two haplotypes identified in our study were from the Artemisia subgenus because they were represented in the same lineage with A. sylvatica, A. argyi and A. verlotiorum. Taken together, the obtained results suggest that rDNA-ITS is not a useful marker for studies concerning the intra-specific molecular diversity of a particular Artemisia species. On the other hand, it could be used in basic research to indicate the proper phylogenetic affiliation of analysed Artemisia plants, because, given its highly conserved nature in Artemisia, it could be supposed that even a single or two substitutions might indicate a unique Artemisia species.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Barney JN, DiTommase A. The biology of Canadian weeds. 118. Artemisia vulgaris L. Can J Plant Sci. 2003;83:205–215.
- Hayat MQ, Ashraf M, Khan MA, et al. Palynological study of the genus Artemisia (Asteraceae) and its systematic simplications. Pak J Bot. 2010;42(2):751–763.
- Miller RA. Mugwort (Artemisia vulgaris L.) [Internet]. Alternative Nature Online Herbal. Cunningham (TN): Alternative Nature Herbals; c1997–2014 [cited 2016 Jan 14]. Available from: http://www.altnature.com/gallery/mugwort.htm.
- Hwang YS, Wu KH, Kumamoto J, et al. Isolation and identification of mosquito's repellents in Artemisia vulgaris. J Chem Ecol. 1985;11(9):1297–1306.
- Chevallier A. The encyclopedia of medicinal plants. London : Dorling Kindersley; 1996.
- Bunyapraphatsara N. Thai medicinal plants used in primary health care system. Bangkok: Medicinal Plant Information Center: Mahidol University; 1986;2:32.
- Wright WC. Artemisia. London: Taylor and Francis; 2002. p. 1–344.
- Vallès J, McArthur ED. Artemisia systematics and phylogeny: cytogenetic and molecular insights. In: McArthur ED, Fairbanks DJ, editors. Shrubland Ecosystem Genetics and Biodiversity. Proceedings; 2000 Jun 13–15; Provo, UT; 2001. p. 67–74.
- Sanz M, Vilatersana R, Hidalgo O, et al. Molecular phylogeny and evolution of floral characters of Artemisia and allies (Anthemideae, Asteraceae): evidence from nrDNA ETS and ITS sequences. Taxon. 2008;57(1):66–78.
- Hobbs CR, Baldwin BG. Asian origin and upslope migration of Hawaiian Artemisia (Compositae–Anthemideae). J Biogeogr. 2013;40:442–454.
- Pellicer J, Hidalgo O, Garnatje T, et al. Life cycle versus systematic placement: phylogenetic and cytogenetic studies in annual Artemisia (Asteraceae, Anthemideae). Turk J Bot. 2014;38:1–8.
- Watson LE, Bates PL, Evans TM, et al. Molecular phylogeny of Subtribe Artemisiinae (Asteraceae), including Artemisia and its allied and segregate genera. BMC Evol Biol. 2002;2(17):1–12.
- McArthur ED, Plummer AP. Biogeography and management of native Western Shrubs: a case study, section Tridentatae of Artemisia. Great Basin Nat Memoirs. 1978;2:229–243.
- McArthur ED, Pope LC. Karyotypes of four Artemisia species: A. carruthii, A. filifolia, A. frigida, and A. spinescens. Great Basin Nat. 1979;39(4):419–426.
- Kornkven AB, Watson LE, Estes JR. Molecular phylogeny of Artemisia section Tridentatae (Asteraceae) based on chloroplast DNA restriction site variation. Syst Bot. 1999;24(1):69–84.
- Lee JH, Lee, JW, Sung JS, et al. Molecular authentication of 21 Korean Artemisia species (Compositae) by polymerase chain reaction-restriction fragment length polymorphism based on trnL-F region of chloroplast DNA. Biol Pharm Bull. 2009;32(11):1912–1916.
- Mahmood T, Hassan N, Nazar N, et al. Phylogenetic analysis of different Artemisia species based on chloroplast gene RPS11. Arch Biol Sci. 2011;63(3):661–665.
- Tripathi KP, Roy S, Maheshwari N, et al. SSR polymorphism in Artemisia annua: recognition of hotspots for dynamics mutation. Plant Omics J. 2009;2(6):228–237.
- Shafie B, Shafie M, Sayed MZH, et al. RAPD and ISSR markers for comparative analysis of genetic diversity in wormwood capillary (Artemisia capillaris) from Negeri Sembilan, Malaysia. J Medicinal Plants Res. 2011;5(18):4426–4437.
- McArthur ED, Mudge J, Van Buren R, et al. Randomly amplified polymorphic DNA analysis (RAPD) of Artemisia subgenus Tridentatae species and hybrids. Great Basin Nat. 1998;58(1):12–27.
- McArthur ED, Van Buren R, Sanderson SC, et al. Taxonomy of Sphaeromeria, Artemisia and Tanacetum (Compositae, Anthemideae) based on randomly on amplified polymorphic DNA (RAPD). Great Basin Nat. 1998;58(1):1–11.
- Sangwan RS, Sangwan NS, Jain DC, et al. RAPD profile based genetic characterization of chemotypic variants of Artemisia annua L. Biochem Mol Biol Int. 1999;47(6):935–944.
- Kurşat M, Civelek Ş, Türkoğlu I, et al. A new species of subgenus Seriphidium of Artemisia L. (Asteraceae) from Turkey. Turk J Bot. 2015;39:88–95.
- Haymes KM. Mini-Prep method suitable for a plant breeding program. Plant Mol. Biol. Rep. 1996;14(3):280–284.
- White TJ, Burns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA gene for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. p. 315–322.
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acid S. 1999;41:95–98.
- Thompson JD, Gibson TJ, Plewniak F, et al. The ClustalX-Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882.
- Akaike H. A new look at statistical model identification. IEEE T Automat Contr. 1974;19:716–723.
- Guindon S, Gascuel O. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704.
- Posada D. jModel test: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256.
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425.
- Eck RV, Dayhoff MO. Evolution of the structure of ferredoxin based on living relics of primitive amino acid sequences. In: Dayhoff MO, editor. Atlas of protein sequence and structure. Silver Spring (MD): National Biomedical Research Foundation; 1966.
- Fitch W. On the problem of discovering the most parsimonious tree. Am Nat. 1977;111:223–257.
- Swofford DL. PAUP* Phylogenetic analysis using parsimony *and other methods. Version 4 beta 10.(MA). Sunderland: Sinauer Associates; 1998.
- Efron B. The jackknife, the bootstrap and other resampling plans: CBMS-NSF MA, Monograph 38, Philadelphia (PA): SIAM; 1982.
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791.
- Tamura K, Stecher G, Peterson D, et al. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30:2725–2729.
- Schlötterer C, Hauser MT, Von Haeseler A, et al. Comparative evolutionary analysis of rDNA ITS regions in Drosophila. Mol Biol Evol. 1994;11(3):513–522.
- Bakker FT, Olsen JL, Stam WT. Evolution of nuclear rDNA ITS sequences in the Cladophora albida/sericea clade (Chlorophyta). J Mol Evol. 1995;40:640–651.
- Kornkven AB, Watson LE, Estes JR. Phylogenetic analysis of Artemisia section Tridentatae (Asteraceae) based on sequences from the internal transcribed spacers (ITS) of nuclear ribosomal DNA. Am J Bot. 1998;85(12):1787–1795.
- Vallès J, Torrell M, Garnatje T, et al. The genus Artemisia and its allies: phylogeny of the subtribe Artemisiinae (Asteraceae, Anthemideae) based on nucleotide sequences of nuclear ribosomal DNA internal transcribed spacers (ITS). Plant Biol. 2003;5(3):274–284.
- Haghighi AR, Belduz AO, Vahed MM, et al. Phylogenetic relationships among Artemisia species based on nuclear ITS and chloroplast psbA-trnH DNA markers. Biologia. 2014;69(7):834–839.
- Kurşat M, Türkoğlu İ, Civelek Ş, et al. A new subspecies record for the flora of Turkey: Artemisia santonicum L. subsp. Patens (Neilr.) K.M. Perss. (Asteraceae). Turk J Bot. 2011;35(1):89.
- Kurşat M, Civelek Ş, Turkoğlu İ, et al. Artemisia sieberi Bess. subsp. Sieberi A new record for Turkey and a delete record for Turkey Artemisia herba-alba Asso. (Asteraceae). Pakistan J Bot. 2011;43:1819–1821.
- Tabur S, Kurşat M, Öney S, et al. New or rare data on chromosome numbers and karyomorphology of some taxa in the subgenus Seriphidium (Bess.) Rouy. (Artemisia, Asteraceae) in Turkey. Caryologia Int J Cytol Cytosystem Cytogenet. 2014;67:305–313.
