ABSTRACT
It has been widely reported that metallothioneins (MTs) play pivotal roles in the metabolism of the relatively nontoxic essential metals, as well as of toxic heavy metals. MT gene polymorphisms in an individual may increase or decrease the expression efficiency of the gene. The present study was aimed to investigate the frequency of genetic variation of four MTs in the Iranian population. Whole blood samples were collected from 300 Iranian healthy individuals. Polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) and automated Sanger sequencing were used for DNA genotype determination. The frequencies of major homozygous, heterozygous and minor homozygous genotypes in MT1 (A>G) were, respectively, 76.0%, 24.0% and 0.0%; in MT1A (C>G), 88.0%, 12.0% and 0.0%; in MT1M (A>C), 86.0%, 14.0% and 0.0%; in MT4 (G>A), 80.0%, 18.7% and 1.3%. The present study showed for the first time that the polymorphism frequency distribution of these four MTs significantly differed between the studied Iranian population and other populations around the world, except for the MT4 polymorphism in the Iranian population and populations from the USA and New Zealand, which were similar. In conclusion, it could be suggested that regional and ethnic differences are the main reasons for this varied prevalence.
Introduction
Metallothioneins (MTs) are highly conserved and low-molecular-weight (6–7 kDa) thiol-rich proteins that bind heavy metals through cysteine residues. This binding protects cells and tissues against the toxicity of heavy metals (Cd, As, Pb and Hg).[Citation1–4] MTs have significant effects in the homeostatic regulation of essential metals such as copper and zinc.[Citation5] In vitro studies show that MTs may participate in cell apoptosis, proliferation and differentiation and as anti-inflammatory mediators.[Citation6–9] Four main MT isoforms (MT1, MT2, MT3 and MT4) are expressed in human organs (especially in the liver and kidneys). The coding sequences are homologous and are located on human chromosome 16 (16q12-22).[Citation10,Citation11]
MT1A missense (A>G) (rs8052394), MT1A 5′ near gene (C>G) (rs9922957), MT1M missense (A>C) (rs1827210) and MT4 missense (G>A) (rs11643815) are non-synonymous single nucleotide polymorphisms (SNPs) located in the coding region of the MT gene. The first one, rs8052394, in exon 3 is due to a nucleotide change of A to G, leading to an amino acid substitution at position 51 (Lys51Arg) of the encoded protein. This change decreases the stability of the protein and its detoxification potential.[Citation12] MT1A (C>G) with an SNP at position Ch16:56672380 is near the 5′ end of the gene with a polymorphic site ‘MRE proximity’.[Citation13] Unlike the other MTs, MT4 is expressed in certain stratified squamous epithelia.[Citation14] Gundacker et al. [Citation15] showed that subjects with the MT4 major homozygote variant GG have lower hair mercury level compared to ones with GA and AA. The rs11643815 SNP frequency distribution has also been analyzed in the US population and showed lower hair mercury.[Citation16]
Whilst there have been more than a few studies investigating the expression of MTs in many diseases, to the best of our knowledge, there have been no studies published to date investigating genetic variants in MTs in Iranian populations. The aim of this study was to identify SNPs in these genes and to genotype those in an Iranian population to examine the results for evidence of genetic association between any of the MT SNPs. Therefore, we aimed to determine and analyze the allele frequencies of four SNPs in MT genes (MT1A missense (A>G), MT1A 5′ near gene (C>G), MT1M missense (A>C) and MT4 missense (G>A)).
Subjects and methods
Subjects
Three hundred normal blood samples were examined from genetically unrelated, healthy volunteers: 150 males with a mean age of 44.47 ± 20.13 years (25–70 years old) and 150 females with a mean age of 43.54 ± 18.33 years (30–70 years old). Samples were collected from an accredited medical diagnostic laboratory in Ahvaz city (southwest of Iran) in the period from October 2014 to December 2014. All blood samples were stored at −20 °C before DNA extraction.
To collect the demographic information, a brief questionnaire was given.
The present study was approved by the Research Ethics Committee of Ahvaz Jundishapur University of Medical Sciences.
Genotyping procedure
Extraction of genomic DNA from 100 µL of whole blood was performed by a Diatom DNA Kit (Bioneer, Korea), according to the method recommended by the manufacturer, and 1% agarose gel electrophoresis (Payapajoohesh, Iran) was used to assess the quality of extracted DNA. Then, extracted DNA samples were stored at −20 °C until further use. First, polymerase chain reaction (PCR) was used to detect the SNPs. PCR was done for each primer () in 25 µL reaction mixture containing 50 ng of genomic DNA, 10 pmol of each primer, 1 U of Taq polymerase (Jena Bioscience, Germany) and 12.5 µL of Master mix (Jena Bioscience, Germany). The PCR conditions were as follows: 4 min at 94 °C for an initial denaturation step; 30 s at 94 °C, 30 s at 62 °C and 45 s at 72 °C for 30 cycles, and finally 7 min at 72 °C for an extension step. Restriction fragment length polymorphism (RFLP) was used for MT1A (rs8052394) genotyping by PstІ restriction enzyme (Vivantis, Malaysia). Electrophoresis of PCR and RFLP products was performed in 1.5% and 2% agarose gels, respectively.
Table 1. List of selected metallothionein SNPs in this study and the primers used for amplification.
DNA sequencing
Automated Sanger sequencing (Applied Biosystems, USA, 2012) and BioEdit software (version 7.2.5) were used for determination of the SNPs in MT1A (rs9922957), MT1M (rs1827210) and MT4 (rs11643815). The information and specifications for each primer are listed in
Statistical analysis
If the genotype and allele frequencies of the polymorphisms fitted the Hardy–Weinberg equilibrium, statistical analysis was done by χ2 analysis. Data analysis was performed using SPSS (version 16) statistical software for Windows. Finally, the results were compared with other population studies and differences were considered as statistically significant at P < 0.05.
Results and discussion
Individual susceptibility plays an important role in the magnitude and the extent of adverse health effects caused by environmental agents.[Citation17] Genetic polymorphisms in common regulatory elements such as metal responsive elements (MREs), glucocorticoid responsive elements (GREs) and antioxidant-responsive elements (AREs) may considerably affect the MT levels.[Citation18,Citation19] Different studies showed that SNPs in MTs can increase the risk of anthropogenic heavy metals (Hg and Cd) toxicity in humans. However, these studies are limited [Citation15,Citation16] and, to the best of our knowledge, there is no study that characterizes these polymorphisms in Iranian groups.
MTs are known to play an important role in maintaining the physiological functions and the SNPs in the genes that encode them show racial differences, leading to different levels of toxicants. These SNPs might also have an effect on cancer development.[Citation20–22] Therefore, determination of the allele frequencies and genotyping of MTs could help to develop regional programmes for reduction of the risk of heavy metal and xenobiotics toxicity. Our results suggest that the Iranian population may be susceptible to heavy metals poisoning. Furthermore, we can reduce the risk of related diseases and cancers.
The frequency of MT1A (A>G), MT1A (C>G), MT1M (A>C) and MT4 (G>A) SNPs was determined by PCR-RFLP () and DNA sequencing (–) in 300 normal subjects in Iran. The statistical analysis (P > 0.05) showed that the data for the four polymorphisms fitted with the Hardy–Weinberg equilibrium. There was no significant difference between the two genders.
Figure 1. Representative PCR-RLFP results for the MT1A (A>G) polymorphism. Lane A, DNA ladder (Vivantis, Malaysia). Lanes B, D, G and O, A/G (heterozygous) genotype; Lanes C, E, F, H, I, J, K, L, M, N and P, A/A (major homozygous) genotype. Note: Agarose gel (2%). PstІ restriction enzyme was used for digestion of the PCR product (743 bp).
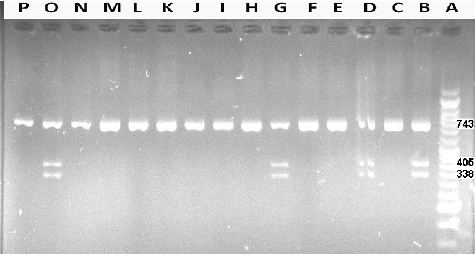
Figure 2. Chromatogram of DNA sequencing for the MT1A (C>G) polymorphism. Major homozygous individual with a C/C genotype. Note: Reverse primer was used. Visualization with BioEdit (v. 7.2.5) software.
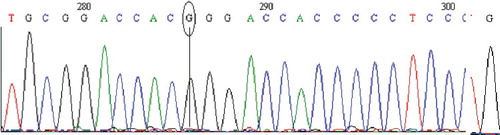
Figure 3. Chromatogram of DNA sequencing for the MT4 (G>A) polymorphism. Minor homozygous individual with an A/A genotype. Note: Reverse primer was used. Visualization with BioEdit (v. 7.2.5) software.
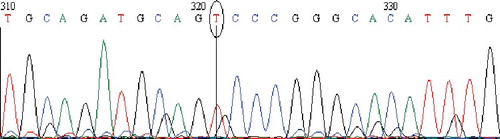
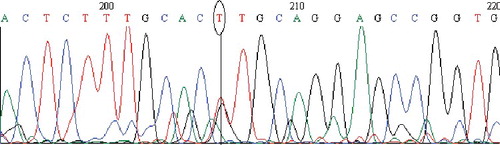
Figure 4. Chromatogram of DNA sequencing for the MT1M (A>C) polymorphism. Heterozygous individual with an A/C genotype. Note: Reverse primer was used. Visualization with BioEdit (v. 7.2.5) software.
MT1A missense (A>G) genotype and allele frequency
Regarding the MT1A (A>G) gene polymorphism, the genotype frequency among the studied individuals was as follows: 76.0% wild type (A/A), 24.0% heterozygote (A/G: three bands of 743, 405 and 338 bp) and 0.0% homozygote (G/G); 88.0% allele A and 12.0% allele G (). The genotype frequency distribution has been studied in Nepalese,[Citation12] West African and North American populations.[Citation23] Yang et al. [Citation24] found that the G allele frequency of MT1A (A>G) is significantly associated with type 2 diabetes (T2DM). In a Japanese population, five polymorphisms were reported to be moderately associated with the risk of lung cancer.[Citation25] In a population from the United States, individuals with the GG and GA genotype of rs8052394 were shown to have lower hair mercury levels than those with the AA genotype. The MT1A (A>G) genotype distribution in Iran differed in comparison to those reported in Europe (P < 0.0001),[Citation26] Spain (P < 0.0001),[Citation24] Italy (P < 0.0001),[Citation21,Citation27] Bulgaria (P < 0.0001),[Citation28] the United States (P < 0.0001),[Citation16] China (P < 0.0001),[Citation24] Nepal (P < 0.0001) [Citation12] and Taiwan (P < 0.0001) [Citation29,Citation30] ().
Table 2. Genotype and allele frequencies of MT1A (A>G), MT1A (C>G), MT4 and MT1M in the studied Iranian population.
Table 3. Comparison between the genotype distribution of MT1A (A>G), MT1A (G>C), MT4 (G>A) and MT1M (A>C) in Iranian population and other populations.
MT1A 5′ near gene (C>G) genotype and allele frequency
The genotype frequency of the MT1 (C>G) polymorphism in wild-type (C/C), heterozygous (C/G) and homozygous (G/G) variants was 88.0%, 12.0% and 0.0%, respectively; with 94.0% allele C and 6.0% allele C (). The studies on the MT1A (C>G) polymorphism are limited. Therefore, the genotype distribution was only compared with available data for the US population [Citation16] (), which showed statistically significant differences (P < 0.0001).
MT1M missense (A>C) genotype and allele frequency
The obtained results about the MT1M (A>C) polymorphism showed that the wild-type (A/A) frequency was 86.0%, that of the heterozygous variant (A/C) was 14.0% and of the homozygous variant (C/C), 0.0%; or 93.0% for allele A and 7.0% for allele C. There are data that MT1M with the ‘Lys20Thr’ polymorphic site has been identified in hepatocellular carcinomas (HCC). HCC cell growth in vitro and in vivo was hampered by modified expression of MT1M.[Citation31] The risk of HCC increased among individuals carrying the rs8052394 A allele.[Citation30] These polymorphisms have been associated partially with metal biomarkers concentrations at levels of exposure relevant to the general population.[Citation32] In this study, when we compared the MT1M (A>C) genotype distribution with NCBI data [Citation23] about other countries, such as China, Japan, Nigeria, the Unites States and European countries, there were significant differences (P < 0.0001) between the Iranian population studied by us and these other populations ().
MT4 missense (G>A) genotype and allele frequency
In our study cohort, the MT4 (G>A) polymorphism frequencies were found to be 80.0% for the wild-type (G/G), 18.7% for the heterozygote (G/A) and 1.3% for the homozygote (A/A) variant; or 89.3% for allele G and 10.7% for allele A (). For the purpose of comparative analysis between our data for the MT4 (G>A) genotype distribution and those available for other populations, mainly data derived from the NCBI website [Citation23] was used. The statistical analysis revealed significant differences between the MT4 (G>A) polymorphism frequencies obtained in our study and those known [Citation24] for Japan (P < 0.0001), China (P < 0.0001), Nigeria (P < 0.0001), Kenya (P < 0.01), Italy (P < 0.0001), Europe (P < 0.0001) and Mexico (P < 0.0001); except two reports, about the United States (P > 0.05) [Citation16] and New Zealand (P = 0.049) populations.[Citation33] In other words, the results from the present study and previous data show that the MT4 (G>A) frequency is significantly different in comparison to that in all other analyzed populations, except the United States and marginally New Zealand ().
Comparison among MT1A (A>G), MT1A (C>G), MT1M (A>C) and MT4 (G>A) genotype frequencies
All of the selected SNPs are located in regulatory regions on autosomal chromosome 16.[Citation16] Therefore, the frequency of the SNPs can be expected to have evolved over time, especially in the Iranian population, which consists of different ethnicities. Because of this diversity, when we compared the genotype and allele frequencies between the two genders, the polymorphism distribution was similar.
The results showed that the prevalence of the wild-type variant of MT1A rs8052394 is lower and that of the heterozygous one is higher compared to the prevalence of the other SNPs analyzed in our study (). Moreover, the frequency percentages of the minor homozygous genotypes of all SNPs in this study were shown to be zero, except that of MT4 rs11643815, which was found to be 1.3% (). However, as our results are based on a cohort of 300 people only, which constitutes a very low per cent of the whole Iranian population, it cannot be excluded that the minor homozygous genotypes of the other three SNPs might possibly also be present in the Iranian population but at a very low frequency.
Additionally, the MTs SNP genotype frequencies studied by us were compared to data about other populations around the world, such as Asian, European, African and American (). Since there is scarce literature about the prevalence of these particular polymorphisms, we referred not only to available articles, but also the NCBI site data.[Citation23] According to the Chi-square test, there was no evidence for genetic association between the four polymorphisms studied (P < 0.0001), except between the MT1A (C>G) and the MT1M (A>C) polymorphisms (P > 0.05), perhaps because they are positioned closer together than the others on chromosome 16. The presence of a minor homozygous genotype only in the MT4 (G>A) polymorphism might possibly be due to its positioning far from the other SNPs on chromosome 16 ().
Taken together, our results and those previously reported demonstrate that there are variations in the MT polymorphisms in East Asia and Europe and Africa or America (). This suggests that ethnicity and geography could be considered as the most important factors in these variations. From a different perspective, the high frequency of the MT major homozygous (wild-type) genotype in our study cohort and in other populations indicates that the MT gene is highly conserved, preserving the MT protein structure.[Citation34]
From a social and public health perspective, it is noteworthy that individuals are different in their response to all materials including chemicals. That is why each polymorphism represents a chance for an individual to be at risk if exposed to a particular chemical pollutant and/or heavy metals. As Iran is a petroleum-exporting and industrial country, its population is more and more exposed to heavy metals. In this context, our data that there was high polymorphism in the MT gene in the studied cohort suggest that a considerable part of the Iranian population may be more sensitive to heavy metals. Thus, more extensive studies would be needed in the future to propose adequate regional programmes and protective measures to reduce the risks of potential toxicity by genetic analysis of exposed people.
Conclusions
To the best of our knowledge, the present study provides the first report on the frequency distribution of four SNPs in the MT gene in the Iranian population. The obtained results suggest that the observed differences between the SNP genotype frequency distributions in the studied cohort and other populations around the world could most likely be attributed to regional and ethnic differences. Based on studies on the MTs SNP genotype frequencies and their distribution, very valuable data and information can be obtained that could help to reveal possible relationships between MTs SNP genotypes and many diseases such as different types of cancer and the effects of toxic substances on the human body. More studies on MTs SNP as rs9922957 need to be done to achieve further success.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgements
The authors would like to thank Mrs Mohaghegh (School of Sciences, Shaheed Chamran University, Ahvaz) and Mr Miah (Toxicology Research Center, Ahvaz Jundishapur University of Medical Sciences, Ahvaz) for their suggestions and guidance in performing this study.
Additional information
Funding
References
- Aschner M, Syversen T, Souza DO, et al. Metallothioneins: mercury species-specific induction and their potential role in attenuating neurotoxicity. Exp Biol Med. 2006;231:1468–1473.
- Buico A, Cassino C, Dondero F, et al. Radical scavenging abilities of fish MT-A and mussel MT-10 metallothionein isoforms: an ESR study. J Inorganic Biochem. 2008;102:921–927.
- Flora S, Mittal M, Mehta A. Heavy metal induced oxidative stress & its possible reversal by chelation therapy. Indian J Med Res. 2008;128:501.
- Carpenè E, Andreani G, Isani G. Metallothionein functions and structural characteristics. J Trace Elem Med Biol. 2007;21:35–39.
- Sigel A, Sigel H, Sigel RK. Metallothioneins and related chelators. Vol. 5. Chicago, IL: Royal Society of Chemistry; 2009.
- Inoue K-i, Takano H, Shimada A, et al. Metallothionein as an anti-inflammatory mediator. Mediators Inflamm. [Internet]. 2009 [cited 2015 Aug 1]; 101659, 7 pages. Available from: http://dx.doi.org/10.1155/2009/101659
- Dutsch-Wicherek M, Sikora J, Tomaszewska R. The possible biological role of metallothionein in apoptosis. Front Biosci. 2007;13:4029–4038.
- Bagheri PM, Govaerts I, De Ley M. Role of metallothionein in differentiation of leukemia cells. Mol Biol Rep. 2011;38:3017–3022.
- Thirumoorthy N, Manisenthil Kumar K, Shyam Sundar A, et al. Metallothionein: an overview. World J Gastroenterol. 2007;13:993–996.
- Li Y, Maret W. Human metallothionein metallomics. J Anal At Spectrom. 2008;23:1055–1062.
- Laukens D, Waeytens A, De Bleser P, et al. Human metallothionein expression under normal and pathological conditions: mechanisms of gene regulation based on in silico promoter analysis. Crit Rev Eukaryot Gene Expr. 2009;19:301–317.
- Timilsina U, Singh K, Tamang H, et al. Polymerase chain reaction-restriction fragment length polymorphism analysis of nsSNP rs8052394 of metallothionein 1A gene. Biomed Res. 2013;24:67–71.
- Hariani G. Application of next generation sequencing technologies to pharmacogenomics [Dissertation]. Raleigh (NC): North Carolina State University; 2012.
- Chen P, Parks WC. Role of matrix metalloproteinases in epithelial migration. J Cellular Biochem. 2009;108:1233–1243.
- Gundacker C, Wittmann KJ, Kukuckova M, et al. Genetic background of lead and mercury metabolism in a group of medical students in Austria. Environ Res. 2009;109:786–796.
- Wang YA Gene-environment study of metallothionein single nucleotide polymorphisms, mercury biomarker levels and peripheral nerve function [dissertation]. Ann Arbor (MI): The University of Michigan; 2011.
- Collins FS, Gray GM, Bucher JR. Transforming environmental health protection. Science. 2008;319:906–907.
- Morahan JM, Yu B, Trent RJ, et al. Genetic susceptibility to environmental toxicants in ALS. Am J Med Genet Neuropsychiat Genet. 2007;144:885–890.
- Raudenska M, Gumulec J, Podlaha O, et al. Metallothionein polymorphisms in pathological processes. Metallomics. 2014;6:55–68.
- McElroy JA, Bryda EC, McKay SD, et al. Genetic variation at a metallothionein 2A promoter single-nucleotide polymorphism in white and black females in Midwestern United States. J Toxicol Environ Health. 2010;73:1283–1287.
- Giacconi R, Bonfigli A, Testa R, et al. +647 A/C and +1245 MT1A polymorphisms in the susceptibility of diabetes mellitus and cardiovascular complications. Mol Genet Metabol. 2008;94:98–104.
- Pedersen MØ, Larsen A, Stoltenberg M, et al. The role of metallothionein in oncogenesis and cancer prognosis. Progress Histochem Cytochem. 2009;44:29–64.
- NCBI: dbSNP Short Genetic Variations [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); c2005 [cited 2016 May 20]. Available from: http://www.ncbi.nlm.nih.gov
- Yang L, Li H, Yu T, et al. Polymorphisms in metallothionein-1 and-2 genes associated with the risk of type 2 diabetes mellitus and its complications. Am J Physiol-Endocrinol Metabol. 2008;294:E987–E992.
- Nakane H, Hirano M, Ito H, et al. Impact of metallothionein gene polymorphisms on the risk of lung cancer in a Japanese population. Mol Carcinogenesis. 2015;54(S1):E122–E128.
- Giacconi R, Simm A, Santos AN, et al. Influence of +1245 A/G MT1A polymorphism on advanced glycation end-products (AGEs) in elderly: effect of zinc supplementation. Genes Nutrition. 2014;9:1–10.
- Cipriano C, Malavolta M, Costarelli L, et al. Polymorphisms in MT1A gene coding region are associated with longevity in Italian Central female population. Biogerontology. 2006;7:357–365.
- Kozarova R, Postadzhiyan A, Apostolova M. Association of +1245 A/G MT1A and-209 A/G MT2A polymorphysms with coronary artery disease and diabetes mellitus in Bulgarian cohort. Biotechnol Biotechnol Equip. 2012;26:100–106.
- Zavras A, Yoon A, Chen M, et al. Metallothionein-1 genotypes in the risk of oral squamous cell carcinoma. Ann Surg Oncol. 2011;18:1478–1483.
- Wong R-H, Huang C-H, Yeh C-B, et al. Effects of metallothionein-1 genetic polymorphism and cigarette smoking on the development of hepatocellular carcinoma. Ann Surg Oncol. 2013;20:2088–2095.
- Mao J, Yu H, Wang C, et al. Metallothionein MT1M is a tumor suppressor of human hepatocellular carcinomas. Carcinogenesis. 2012;33:2568–2577.
- Wang Y, Goodrich JM, Gillespie B, et al. An investigation of modifying effects of metallothionein single-nucleotide polymorphisms on the association between mercury exposure and biomarker levels. Environ Health Perspect. 2012;120:530–534.
- Morgan AR, Fraser AG, Ferguson LR. Metallothionein genes: no association with Crohn's disease in a New Zealand population. J Negat Results Biomed. 2012;11:34–38.
- Chatterjee M. A review of metallothionein isoforms and their role in pathophysiology. World J Surg Oncol [ Internet]. 2011 [cited 2015 Aug 14]; 20;9:54. Available from: http://dx.doi.org/10.1186/1477-7819-9-54
