ABSTRACT
MicroRNAs (miRNAs) binding to the 3′-untranslated regions (3′UTRs) of messenger RNAs to mediate translation and thereby regulate cell differentiation, apoptosis and tumorigenesis. Single nucleotide polymorphisms (SNPs) in the 3′UTRs of genes would alter the binding affinity between targeted genes and miRNA so as to change individual gene expression. We genotyped miRNA-binding-site SNPs in a case-control study with sporadic colorectal cancer (CRC) patients to identify CRC risk associated SNPs. Five miRNA-binding-site SNPs located in the 3′UTR of KRT81 (rs3660), RYR3 (rs1044129), KIAA0423 (rs1053667), C14orf101 (rs4901706) and GOLGA7 (rs11337) were genotyped in CRC patients, using polymerase chain reaction–ligase detection reaction (PCR–LDR) methods. Renilla luciferase reporter assays were used to measure the binding affinity between miRNA and the targeted gene. The χ2 test was used to study the association between carriership of miRNA-binding-site SNPs and CRC risk. The SNP of rs1053667 located in the 3’UTR of KIAA0423 gene was identified to be associated to CRC cancer risk, with the TT allele being associated with a 2.030-fold increased risk for CRC than that associated with the C/T+C/C genotype (odds ratio of 2.030; 95% confidence interval of 1.314–3.136; P < 0.01). The reduced Renilla luciferase activity of rs1053667 TT type comparing with that of CC genotype also demonstrated the different binding affinity of rs1053667 SNPs to the corresponding miRNA. The miRNA-binding-site SNP of rs1053667 in the KIAA0423 gene could be associated with risk of CRC.
KEYWORDS:
Introduction
Colorectal cancer (CRC) is the third most common cancer and is responsible for about 700,000 deaths each year, which makes it the fourth leading cause of cancer-related death in both sexes worldwide.[Citation1] Accumulating evidence that the incidence of the disease is significantly increasing in most developing countries suggests that the disease burden could be expected to become even heavier in the near future.[Citation2,Citation3] Sporadic colorectal cancer is the most common type of CRC that occurs in older population, in the absence of family history, usually as an isolated colonic or rectal lesion. CRC risk is considered to be affected by environmental factors, dietary patterns (high fat consumption), genetic and epigenetic changes.[Citation4–7]
MicroRNAs (miRNAs) are highly conserved small non-coding RNAs with lengths of ∼22 nucleotides that function in post-transcriptional regulation of mRNA expression.[Citation8–10] This usually results in gene-silencing via translational repression or target degradation through 2–8 nucleotides known as ‘seed region’ in miRNA by base-pairing with a complementary sequence in the 3′untranslated region (3′UTR) of the target mRNA. [Citation11–13] miRNAs have been implicated in cell proliferation, differentiation, tumorigenesis and development, hormone secretion and apoptosis by regulating the expression of targeted genes.[Citation8,Citation10,Citation14–17]
Single nucleotide polymorphisms (SNPs) of the ‘seed region’ can influence the complementarity between the miRNA and its target mRNA so as to alter the target gene's expression,[Citation18] and a few studies have focused on the relationship of these SNPs in the miRNA-binding site and cancer risk. Landi et al. [Citation19] found statistically significant associations between CRC risk and the miRNA-binding-site SNPs in CD86 and INSR genes. Yu et al. [Citation20] identified 12 cancer risk associated SNPs located in miRNA-targeting sites in a study including many types of cancers as a whole research object. In this study, we genotyped these 12 SNPs in a case-control study with CRC patients from the Hebei area. Six SNPs with minor allele frequencies of less than 5% were excluded, the relationship of SET8 (rs16917496) and CRC is subject of another investigation by a team of collaborators, which is why the remaining five miRNA-binding-site SNPs located in the 3′UTR of KRT81 (rs3660), RYR3 (rs1044129), C14orf101 (rs4901706), KIAA0423 (rs1053667) and GOLGA7 (rs11337) were assessed in the present study in relation to cancer risk.
Materials and methods
Tissue specimens
Blood samples were collected at the Fourth Hospital of Hebei Medical University from 237 patients with CRC who underwent tumour resection in the Department of Surgery and 293 age-matched healthy controls without any history of tumours. A Wizard Genomic DNA extraction kit (Promega, Madison, WI, USA) was used for genomic DNA extraction. All procedures were supervised and approved by the hospital's Human Tissue Research Committee. All the patients signed a written informed consent form for the sample collection procedure and subsequent analysis.
SNP genotyping of miRNA-binding-site SNPs
The miRNA-binding-site SNPs, including KRT81 (rs3660), RYR3 (rs1044129), C14orf101 (rs4901706), KIAA0423 (rs1053667) and GOLGA7 (rs11337) were genotyped using the polymerase chain reaction–ligase detection reaction (PCR-LDR) assay with forward and reverse primers to amplify the DNA fragments flanking the SNPs based on the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/snp/). A PCR Master Mix Kit (Promega, Madison, WI, USA) was used for PCR amplification, and PCR products were used as template for ligation with different probes of different SNPs, and the ligated products were separated using the ABI PRISM Genetic Analyzer 3730XL (Thermo Fisher Scientific, Waltham, MA, USA). All the sequences of the primers and probes are listed in
Table 1. Primer sequences and probes for five SNPs located in miRNA-targeting sites.
Renilla luciferase reporter assay
Four oligonucleotides containing, from 5′ to 3′: XhoI sticky end (5 bp), a fragment from the 3′UTR of the KIAA0423 gene containing the GG or AA genotype (rs1053667; 51 bp) and a NotI sticky end (2 bp), were synthesized: sense for CC (5′-TCGAGATATTTTTGAGAAAAGTCCTGCTCACTTGCACTATTCTATAGAAACTACAAGC-3′) and antisense for CC (5′-GGCCGCTTGTAGTTTCTATAGAATAGTGCAAGTGAGCAGGACTTTTCTCAAAAATATC-3′); sense for TT (5′-TCGAGATATTTTTGAGAAAAGTCCTGCTCATTTGCACTATTCTATAGAAACTACAAGC-3′) and antisense for TT (5′-GGCCGCTTGTAGTTTCTATAGAATAGTGCAAATGAGCAGGACTTTTCTCAAAAATATC-3′). The four oligonucleotides were first denatured with 1 × NEBuffer 2 (New England Biolabs, Ipswich, MA, USA) in a heating block at 95 °C for 5 min, followed by a gradual reduction of temperature to room temperature. The psiCheck2 vector (Promega, Madison, WI, USA) containing Renilla luciferase and controlled firefly luciferase genes was linearized by digestion with NotI and XhoI (New England Biolabs, Ipswich, MA, USA) and purified. The annealed oligonucleotides were ligated in the linearized psiCheck2 vector into the NotI- and XhoI-cloning sites located downstream of the Renilla luciferase reporter gene with T4 DNA ligase (Promega, Madison, WI, USA). The ligated vectors were transformed in Escherichia coli competent cells, and positive clones were selected by sequencing.
The HeLa cell line was seeded in 48-well plates and transfected with 800 ng of the modified psiCheck2 vector containing either the CC or TT genotype. Then, the Renilla luciferase activity was measured with a luminometer (Lumat, Albuquerque, NM, USA) 48 h after transfection with the Dual-Lucy Assay Kit (Vigorous Instrument, Beijing, China) and the transfection efficiency was normalized with the firefly luciferase activity.
Statistical analysis
The χ2 test was used to study the association between carrierships of miRNA-binding-site SNPs and CRC risk in CRC patients and healthy controls. The χ2 test was also used to compare age and gender frequency distribution in the cases and control groups. The odds ratio (OR) and 95% confidence interval (95% CI) were calculated using an unconditional logistic regression model. The t-test was used to compare differential expression levels between genotypic groups with Renilla luciferase reporter assays. All of the statistical analyses were performed using the SPSS 18.0 software package (SPSS Company, Chicago, IL, USA). Differences were considered statistically significant at P < 0.05.
Results and discussion
A total of 237 CRC patients and 293 controls from Shijiazhuang and surrounding areas in North China were enrolled in this study. The clinical factors of the CRC patients and healthy controls are listed in . The distribution frequency in terms of age and gender was not different between the groups of patients and controls.
Table 2. Clinical factors of colorectal cancer patients and controls.
The miRNA-binding-site SNPs including KRT81 (rs3660), RYR3 (rs1044129), C14orf101 (rs4901706), KIAA0423 (rs1053667) and GOLGA7 (rs11337) were genotyped in CRC patients and healthy controls. The genotype distribution frequencies of the SNPs are shown in , all of them fit the Hardy–Weinberg equilibrium in the controls. For rs1053667 in KIAA0423, the genotype distribution frequency of T/T and C/T+C/C was significantly different between CRC patients and healthy controls (OR: 2.030, 95% CI: 1.314–3.136, P < 0.05). The T/T genotype of rs1053667 was associated with a 2.030-fold increased risk when compared with that for the C/T+C/C genotype. We subsequently performed stratified analysis and the cancer risk association of this SNP was also seen in different age groups and different gender groups ().
Table 3. Associations of the five SNPs with colorectal cancer risk.
Table 4. Stratified analysis of the association of rs1053667 with CRC referring to age and gender.
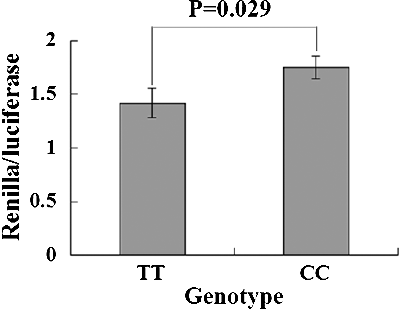
When the Renilla luciferase reporter containing the CC or TT genotype of rs1053667 was constructed and transfected in HeLa cells to measure the binding affinity between the rs1053667 SNP and miRNA, a significant reduction of Renilla luciferase activity was observed in the TT genotype compared with that of the CC type (). These results indicated that the TT type of the rs1053667 SNP in 3′UTR of KIAA0423 had higher binding affinity with the corresponding miRNA. This change in the binding affinity between miRNAs and KIAA0423 could mediate the gene expression so as to modify the risk of CRC.
Figure 1. Renilla luciferase assay for TT and CC genotypes of rs1053667 in HeLa cell lines. The Renilla luciferase activities are defined as the ratio of Renilla luciferase activities versus firefly luciferase activities.
Pathways involving miRNAs have emerged as a crucial system for the regulation of tumorigenesis.[Citation21–24] The miR-SNPs, defined as both SNPs within the miRNA binding site and SNPs in miRNAs themselves, are key factors that modify disease phenotypes.[Citation25,Citation26] We and other researchers have identified a number of cancer risk and outcome associated miR-SNPs in previous studies,[Citation27–31] all of which demonstrated the important role of miRNA regulation in carcinogenesis and progress of cancer. In this study, the miR-SNPs including KRT81 (rs3660), RYR3 (rs1044129), C14orf101 (rs4901706), KIAA0423 (rs1053667) and GOLGA7 (rs11337) were assessed for their association with CRC risk, and rs1053667 in the 3′UTR of KIAA0423 was identified to be potentially associated with CRC risk based on χ2 analysis.
The true mechanism by which rs1053667 modifies the CRC risk remains unclear. As this SNP is located in the 3′UTR of KIAA0423, the SNP might affect mRNA stability and alter the expression of KIAA0423. The Renilla luciferase reporter assay supported our hypothesis that the T-to-C transition of rs1053667 could change the binding affinity between KIAA0423 and corresponding RNAs, thereby modulating KIAA0423 expression. The altered expression of KIAA0423, in turn, might modify the carcinogenesis process associated with CRC. The ‘KIAA’ genes refer to more than 2000 genes isolated mainly from cDNA libraries of brain tissue. The average length of the cDNA for these genes is 4.8 kb and 866 amino acid residues.[Citation32,Citation33] Deletions of some of these KIAA genes seem to be associated with dysfunction of the nervous system.[Citation34] Since a KIAA member has been shown to display association with cancer,[Citation35] functional analysis of KIAA0423 should be performed in further studies to uncover the potential role of this gene in CRC carcinogenesis.
Conclusions
The results from this study suggest that the SNPs in KIAA0423 could be used as potential tumour markers for early cancer detection and prevention. However, our results need further validation in a larger sample size and laboratory based functional study.
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- GLOBOCAN . Estimated cancer incidence, mortality and prevalence worldwide in 2012 [Internet]. Lyon : International Agency for Research on Cancer (IARC); 2012, c2016 [2014 Jan 9], Available from: http://globocan.iarc.fr.
- Sung JJ , Lau JY , Goh KL , et al. Increasing incidence of colorectal cancer in Asia: implications for screening. Lancet Oncol. 2005;6:871–876.
- Bishehsari F , Mahdavinia M , Vacca M , et al. Epidemiological transition of colorectal cancer in developing countries: environmental factors, molecular pathways, and opportunities for prevention. World J Gastroenterol. 2014;20:6055–6072.
- Norat T, Chan D, Lau R, et al. WCRF/AICR Systematic literature review - continuous update project report. The associations between food, nutrition and physical activity and the risk of Colorectal Cancer. World Cancer Research Fund/American Institute for Cancer Research; 2010. Available from: http://www.wcrf.org/sites/default/files/SLR_colorectal_cancer_2010.pdf.
- Migliore L , Migheli F , Spisni R , et al. Genetics, cytogenetics and epigenetics of colorectal cancer. J Biomed Biotechnol [Internet]. 2011 [cited 2016 Jan 4];2011:792362. Available from: http://www.hindawi.com/journals/bmri/2011/792362/.
- Peters U , Jiao S , Schumacher FR , et al. Identification of genetic susceptibility loci for colorectal tumors in a genome-wide meta-analysis. Gastroenterology. 2013;144:799–807.
- Markowitz SD , Bertagnolli MM . Molecular origins of cancer: molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–2460.
- Bartel DP , MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297.
- Cui C , Yu J , Huang S , et al. Transcriptional regulation of gene expression by microRNAs as endogenous decoys of transcription factors. Cell Physiol Biochem. 2014;33:1698–1714.
- Ambros V . The functions of animal microRNAs. Nature. 2004;431:350–355.
- Meister G , Tuschl T . Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349.
- Zeng Y , Wagner EJ , Cullen BR . Both natural and designed microRNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell. 2002;9:1327–1333.
- Long D , Lee R , Williams P , et al. Potent effect of target structure on microRNA function. Nat Struct Mol Biol. 2007;14:287–294.
- Lim LP , Lau NC , Weinstein EG , et al. The microRNAs of Caenorhabditis elegans . Genes Dev. 2003;17:991–1008.
- Lim LP , Glasner ME , Yekta S , et al. Vertebrate microRNA genes. Science. 2003;299:1540.
- He L , Thomson JM , Hemann MT , et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833.
- Iorio MV , Croce CM . MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27:5848–5856.
- Ryan BM , Robles AI , Harris CC . Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389–402.
- Landi D , Gemignani F , Naccarati A , et al. Polymorphisms within micro-RNA-binding sites and risk of sporadic colorectal cancer. Carcinogenesis. 2008;29:579–584.
- Yu Z , Li Z , Jolicoeur N , et al. Aberrant allele frequencies of the SNPs located in microRNA target sites are potentially associated with human cancers. Nucleic Acids Res. 2007;35:4535–4541.
- Arora A , Singh S , Bhatt AN , et al. Interplay between metabolism and oncogenic process: role of microRNAs. Transl Oncogenomics. 2015;7:11–27.
- Yin Y , Song M , Gu B , et al. Systematic analysis of key miRNAs and related signaling pathways in colorectal tumorigenesis. Gene. 2016;578:177–184.
- Zhang QJ , Xu C . The role of microRNAs in the pathogenesis of pituitary tumors. Front Biosci (Landmark Ed). 2016;21:1–7.
- Navarro A , Muñoz C , Gaya A , et al. MiR-SNPs as markers of toxicity and clinical outcome in Hodgkin lymphoma patients. PLoS One [Internet]. 2013 [cited 2013 May 21];8:e64716. Available from: http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0064716.
- Horikawa Y , Wood CG , Yang H , et al. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin Cancer Res. 2008;14:7956–7962.
- Hu Z , Chen J , Tian T , et al. Genetic variants of miRNA equences and non-small cell lung cancer survival. J Clin Invest. 2008;118:2600–2608.
- Peng C , Guo Z , Wu X , et al. A polymorphism at the microRNA binding site in the 3' untranslated region of RYR3 is associated with outcome in hepatocellular carcinoma. Onco Targets Ther. 2015;8:2075–2079.
- Yang B , Liu C , Diao L , et al. A polymorphism at the microRNA binding site in the 3′ untranslated region of C14orf101 is associated with non-Hodgkin lymphoma overall survival. Cancer Genet. 2014;207:141–146.
- Xie Y , Diao L , Zhang L , et al. A miR-SNP of the KRT81 gene is associated with the prognosis of non-Hodgkin's lymphoma. Gene. 2014;539:198–202.
- de Larrea CF , Navarro A , Tejero R , et al. Impact of MiRSNPs on survival and progression in patients with multiple myeloma undergoing autologous stem cell transplantation. Clin Cancer Res. 2012;18:3697–3704.
- Zhao Y , Du Y , Zhao S , et al. Single-nucleotide polymorphisms of microRNA processing machinery genes and risk of colorectal cancer. Onco Targets Ther. 2015;8:421–425.
- Imai K , Kawai M , Tada M , et al. Temporal change in mKIAA gene expression during the early stage of retinoic acid-induced neurite outgrowth. Gene. 2005;364:114–122.
- Nakajima D , Okazaki N , Yamakawa H , et al. Construction of expression-ready cDNA clones for KIAA genes: manual curation of 330 KIAA cDNA clones. DNA Res. 2002;9:99–106.
- Nakayama M , Iida M , Koseki H , et al. A gene-targeting approach for functional characterization of KIAA genes encoding extremely large proteins. FASEB J. 2006;20:1718–1720.
- Schaffrik M , Mack B , Matthias C , et al. Molecular characterization of the tumor-associated antigen AAA-TOB3. Cell Mol Life Sci. 2006;63:2162–2174.
