ABSTRACT
Alpha-lactalbumin (α-LA) is a distinctive whey protein found in bovine milk and the milk of other mammalian species. Its main function in the lactating mammary glands is involved in lactose biosynthesis, which makes it a potential quantitative trait locus. The aim of this study is to determine the incidence of single nucleotide polymorphisms (SNPs) within the exonic and intronic portions of the caprine α-LA gene (LALBA) in native Saudi breeds (Ardi, Habsi and Harri) in relation to utility traits. Blood samples were collected from 300 animals (100 for each breed). Genomic DNA was extracted and a 268 bp fragment of the LALBA gene comprising exon 3 and its flanking area was amplified. Subsequent digestion with MvaI restriction endonuclease resulted in two different banding patterns: αLA A1/αLA A1 and αLA A1/αLA A2, and two allelic forms, i.e. αLA A1 and αLA A2. Nucleotide sequencing of the designated LALBA amplicons was done and, following successful BLAST analysis, the sequences were submitted to GenBank (NCBI Accession No. KM267632, KM267633, KM267634 and KP940442). SNPs were searched within breeds of Saudi origin and homology was sought among caprine, ovine and bovine species. A C > T transition was identified within the fifth nucleotide of the silent α-LA A2 allele. Phylogenetic analysis on the basis of LALBA nucleotide sequence of Saudi goats indicated similarity with evolutionarily related sheep, while the origin of bovine species and deer was located some distance away.
Introduction
The domestic goat (Capra hircus), being one of the earliest livestock species to be domesticated, is still essential in animal husbandry today. Goats are extensively reared for meat, milk, fibre and pelts, especially in the developing countries [Citation1,Citation2].
Alpha-lactalbumin (α-LA) is a distinctive whey protein found in bovine milk and the milk of other mammalian species. Its main role in the lactating mammary glands is involved in regulation of the lactose synthase complex, which makes it a potential quantitative trait locus [Citation3,Citation4]. For review, see [Citation5].
Single nucleotide polymorphisms (SNPs) can affect the amino-acid sequence or the post-translational modifications of milk proteins and their bioactivity [Citation6–7]. The caprine LALBA gene has been less frequently studied than the goat casein genes and their polymorphisms due to limited economic interests [Citation8–11]. Initial reports regarding various LALBA forms emerged in the Italian population [Citation11] and later, in Chinese breeds, different variants were analysed for influence on milk and body size traits [Citation4]. On a global scale, polymorphism has been identified at position 3E+5 and 3E+7 in exon 3 of the LALBA gene [Citation12,Citation13].
Three distinct Saudi Arabian goat breeds of particular economic importance in the region are Ardi, Habsi and Harri. The aim of this study was to evaluate the genetic variability within the LALBA nucleotide sequence, using restriction fragment length polymorphism (RFLP) and non-radioactive single-strand conformation polymorphism (SSCP), followed by Sanger sequencing. This generated baseline data of gene variants (SNPs and INDELS) for utilization in future association studies to establish a breeding programme based on marker-assisted selection (MAS) and to enhance the productivity of the goat genetic resources of Saudi Arabia.
Materials and methods
Experimental animals
Blood samples from Ardi and Harri goats (n = 100 for each breed) were collected from private farms located in the suburbs of Jeddah and Riyadh, while samples belonging to the Habsi breed (n = 100) were obtained from two farms located near Al-Qunfudhah village, South Jeddah. All animals were female goats in their first to second season of lactation with an average age of 1 to 1.5 years.
Milk yield and statistical analysis
Animals were raised under extensive husbandry conditions and were fed on hay (ad libitum) and concentrated feed mixture (14% CP) according to their requirements. Daily records of milk yield were initiated four days after kidding. Milk yield was recorded three days per week during the first 16 weeks of lactation and the average daily and weekly yield were then estimated. The total milk yield for each studied breed was statistically analysed by one-way analysis of variance (ANOVA) and variations were tested by the F-test. We considered p-values of less than 0.05 to indicate statistical significance. Statistical analyses were done with SPSS version 16.0 for Windows.
The genotypes were determined by direct counting of restriction fragments observed in the gel. A standard procedure was employed to calculate the frequency of different genotypes [Citation14]:
Genomic frequencey = n of individuals of a particular genotype/total n of individuals of all genotypes
Blood collection and DNA isolation
About 10 mL of blood was collected from each goat, using 0.5 mL ethylenediaminetetraacetic acid (EDTA; 0.5 mol/L, pH 8) as an anticoagulant. The samples were transported to the animal genetics laboratory and kept at −20 °C until isolation of genomic DNA. Genomic DNA was extracted via the QIAamp DNA extraction Mini Kit (Qiagen).
Polymerase chain reaction (PCR)-RFLP
A 268-bp region spanning over exon 3 and a part of its flanking portion was amplified by employing a set of forward (5’-TCATCTAAAAGGCAACAGGTA-3’) and reverse (5’-ATAGTGCTGGGGCGAAA-3’) primers [Citation15]. Amplifications were carried out in 200 μL PCR tubes and the reaction mixture contained: 5 μL of master mix (5 × Taq Master: DNA polymerase, deoxynucleoside triphosphates (dNTPs), (NH4)2SO4, MgCl2, Tween-20, Nonidet P-40 and stabilizers; Jena Bioscience, Jena, Germany), forward and reverse primers (1 μL each), 2 μL of genomic DNA and PCR-grade water (filled up to 25 μL). The cycling programme (an initial cycle at 95 °C for 5 min followed by 35 cycles of steps containing denaturation at 95 °C for 30 s, annealing at 62 °C for 30 s, extension at 72 °C for 30 s and final extension at 72 °C for 5 min) was performed in a pre-programmed thermal cycler (MULTIGENE, Labnet International Inc., Edison, NJ, United States). Amplified products were visualized through agarose gel electrophoresis (3%) and the bands were stained with ethidium bromide (Bio-Rad, Mississauga, Ontario, Canada).
PCR amplicons were digested for 5 h at 37 °C with 15 U of MvaI restriction endonuclease (Jena Bioscience, Jena, Germany). Restriction enzyme, 10 × reaction-buffer and autoclaved triple distilled water were mixed for all the mandatory reactions in 1.5 mL micro-centrifuge tubes. Following digestion, the samples were resolved in a 4% (w/v) agarose gel in 1 × Tris-borate-EDTA (TBE) buffer stained with ethidium bromide (Bio-Rad, Mississauga, Ontario, Canada) to distinguish between the genotypes.
PCR-SSCP
Aliquots of 10 μL PCR products were blended with 10 μL denaturing solution (95% formamide, 25 mmol/L EDTA, 0.025% xylene-cyanole and 0.025% bromophenol blue), heated for 10 min at 98 °C and chilled on ice [Citation16]. Denatured DNA was subjected to 12% polyacrylamide gel electrophoresis (PAGE) in SCIE-PLAS vertical electrophoresis unit (Cambridge, UK) using 1 × TBE buffer for 14 h at a constant temperature of 10 °C. To visualize the test material in the gel, silver staining was performed. The staining procedure included soaking of the gel for 4 min at room temperature in dark conditions in a 500 mL solution of 0.2% AgNO3, 2% HNO3 and 10% ethanol. Next, the gel was quickly (5 s) rinsed once with 500 mL of distilled H2O and was then developed by soaking at room temperature in a 500 mL developing solution (3% NaOH, 4 mL of 37% HCOH and 0.005% eriochrome black T) until the bands appeared with a sufficient intensity (3 min). Later, the gel was washed simply with tap water to remove the reaction precipitate of silver nitrate. DNA bands were visualized directly in a white light box, then photographed or scanned. The gel plate was agitated in a shaker throughout the staining process [Citation17].
Nucleotide sequencing and sequence analysis
Selected LALBA fragments were sequenced on both strands following the Sanger's dideoxy chain termination method [Citation18]. After successful Basic Local Alignment Search Tool (BLAST) analysis (http://www.ncbi.nlm.nih.gov/BLAST/), the sequences belonging to different alleles were analysed via the ClustalW sub-programme of the BioEdit Sequence Alignment Editor (BioEdit v. 7.2.5, 2013) to generate sequence alignment reports. Phylogenetic analysis was done using the Maximum Likelihood method packaged within the Molecular Evolutionary Genetics Analysis (MEGA v 6.0) tools.
Results and discussion
PCR-RFLP and SSCP analysis
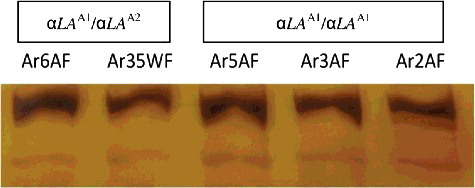
The 268 bp LALBA amplicons were restricted with MvaI to establish polymorphic sites within the coding region of the gene [Citation12]. MvaI, with a recognition site of 5'-CC↓WGG-3'/3'-GGW↑CC-5', revealed two allelic forms (α-LA A1 and α-LA A2) and generated two different banding patterns, or genotypes, i.e. α-LA A1/α-LA A1 and α-LA A1/α-LA A2. The α-LA A1/α-LA A1 genotype was deduced to be normal homozygous wild type, whereas the presence of a silent α-LA A2 allele generated a heterozygous variant (α-LA A1/α-LA A2). The α-LA A1 allele, with a single MvaI site, generated two bands of 140 bp and 128 bp. The α-LA A2 allele, in the absence of a restriction site, produced an undigested product of 268 bp ().
Figure 1. RFLP electrophoretic patterns of LALBA amplicons generated by MvaI restriction endonuclease. Lane 1, 50 bp ladder (Jena Bioscience, Jena, Germany); Lanes 2–10, 12 Habsi, 20 Habsi, 2 Habsi, 30 Ardi, 11 Harri, 4 Harri, 43 Ardi, 24 Ardi and 6 Ardi.
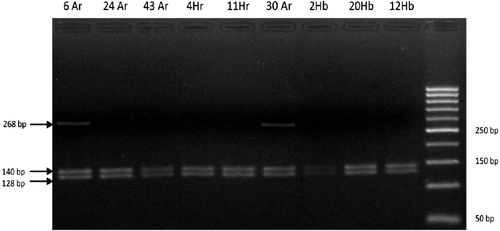
The distribution of the genotypic and allelic frequencies is presented in . The frequency of α- LA A2 allele was found to be lower compared to that of the α-LA A1 allele across the studied breeds. The distribution patterns of LALBA genotypes revealed 98% of all the investigated animals to be homozygous α-LA A1/α-LA A1 and only 2%, heterozygous α-LA A1/α-LA A2. These figures demonstrated the prevalence of the α-LA A1 allele in Saudi goats.
Table 1. Genotype and allele frequencies based on MvaI restriction enzyme digestion of the amplified LALBA fragments and estimated milk yields for the studied goat breeds.
The SSCP gel patterns did not reveal any distinct variation (). As PCR-RFLP already demonstrated the existence of a polymorphic site, designated amplicons were selected for nucleotide sequencing.
Association between LALBA allelic variants and the milk production trait
Earlier studies have not demonstrated significant association between LALBA variation and milk lactose levels [Citation19]. There are reports that suggest that some LALBA variants affect the renneting properties of milk [Citation20]. The results from the linkage analysis between the milk production trait and different LALBA forms are shown in . Based on these data, no significant association was established between the prevalent (αLA A1) or alternative allele (αLA A2) and milk yield (P > 0.05). These results are consistent with the records about Chinese goats [Citation4], Chinese Holstein cows [Citation21] and Indian buffalo [Citation22].
Nucleotide sequence comparison
The nucleotide sequences generated in this study were deposited in GenBank NCBI under accession numbers KM267632, KM267633, KM267634 and KP940442. The BLAST analysis of the LALBA sequences of Saudi goats (NCBI Accession No. KM267632, KM267633 and KM267634) revealed complete similarity with Chinese breeds (NCBI Accession No. DQ629104, DQ629105, DQ629106, DQ629107, DQ629108, DQ629109, DQ629110, DQ629111 and DQ629112).
Comparison of the nucleotide sequences from the present study (NCBI Accession No. KM267632 and KP940442) with that of the already published sequence of the caprine LALBA gene (NCBI Accession No. M63868) via ClustalW revealed complete similarity except for an SNP within the silent allele (αLA A2) (NCBI Accession No. KP940442), i.e. a C > T transition at position 127 and within the fifth nucleotide of exon 3 (TTCPhe > TTTPhe at position 80 of the mature protein) as described earlier [Citation12]. The nucleotide sequence of the variant form showed an ambiguous code ‘Y’ (C or T) that altered the enzyme restriction site ((A,B)). Differences were identified, at the nucleotide sequence level, while aligning the caprine (NCBI Accession No. M63868, KM267632 and KP940442), ovine (NCBI Accession No. AB052168) and bovine LALBA nucleotide sequences (NCBI Accession No. X06366). The ovine and bovine sequences (NCBI Accession No. AB052168 and X06366) showed deletions at positions 101 and 102. Moreover, at positions 20, 54, 120, 155, 162 and 257, the caprine LALBA sequences (NCBI Accession No. M63868 and KM267632) were different from the ovine one (NCBI Accession No. AB052168) and, at positions 4, 6, 8, 20, 67, 108, 110, 111, 112, 155, 162, 234 and 257, from the bovine sequence (NCBI Accession No. X06366) (). These insertions/deletions could be utilized as markers for species differentiation/identification.
Figure 3. (A) Chromatograms showing disparities at the sequence level between αLA A1 and αLA A2 alleles. (B) MvaI restriction sequence within the respective alleles.
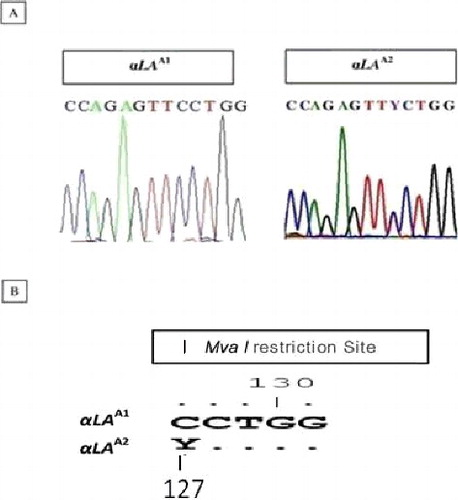
Figure 4. Multiple sequence alignment of exon 3 and flanking region of the goat LALBA gene (NCBI Accession No. KM267632 and KP940442) under study with that of already published sequences of goat (NCBI Accession No. M63868), sheep (NCBI Accession No. AB052168) and Bovine LALBA sequence (NCBI Accession No. X06366).

Phylogenetic analysis
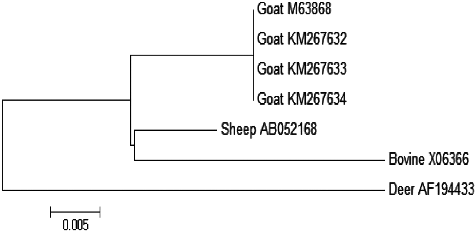
The sequence data of the goat LALBA fragments were compared with already published α-LA sequences of goat (NCBI Accession No. M63868), sheep (NCBI Acc. No. AB052168) and bovine species (NCBI Accession No X06366). The phylogenetic analysis on the basis of the LALBA nucleotide sequence of native Saudi goats (NCBI Accession No. KM267632, KM267633 and KM267634) indicated complete similarity with the reference LALBA sequence of goat (NCBI Accession No. M63868) and close relationship to sheep (NCBI Accession No. AB052168). However, the origin of bovine and deer was located some distance away ().
Figure 5. Phylogenetic analysis based on nucleotide sequences of the LALBA fragments of Saudi goats (NCBI Accession No. KM267632, KM267633 and KM267634) with GenBank α-LA reference sequences of goat (NCBI Accession No. M63868), Ovis aries (NCBI Accession No. AB052168), Bovine (NCBI Accession No. X06366) and deer (NCBI Accession No. AF194433).
The present study was aimed at characterizing polymorphisms of the LALBA gene in Saudi goats and investigating possible associations between LALBA allelic variants and milk yield. It represented a continuation of our earlier work on distinct beta-lactoglobulin gene (LGB) allelic variants significantly associated to high milk yield [Citation23]. The lack of statistically significant association between the prevalent (αLA A1) or alternative allele (αLA A2) and milk yield, might possibly be due to the limited sample size of the resource population. However, it should be taken into account that various molecular indicators within individual populations considerably minimize the variance attributable to aspects linked to that particular environment.
With the advancements in DNA analysis technology, individual mutations which are electrophoretically impossible to differentiate can be identified in both the coding and non-coding regions. This allows easy and prompt identification of carriers of favourable alleles and speeds up genetic improvement. Our future efforts will aim at a deeper evaluation of SNPs in the native caprine species and establishment of a MAS-based breeding programme.
Conclusions
This study showed that neither the wild-type αLA A1 allele, nor the silent α-LA A2 allele with the identified C > T transition in the fifth nucleotide could be considered as significantly associated to milk yield in Ardi, Habsi and Harri goats. The obtained nucleotide sequences enlarged the catalogue of caprine LALBA genotypic data and increased the panel of identified SNPs. The results from this study provide a useful basis towards characterization of SNPs within the indigenous goat breeds of Saudi Arabia.
Acknowledgments
The authors acknowledge the assistance of the Science & Technology Unit, Deanship of Scientific Research and Deanship of Graduate Studies, King Abdulaziz University, Jeddah, KSA.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Zeder MA , Hesse B . The initial domestication of goats (Capra hircus) in the Zagros Mountains 10,000 years ago. Science 2000;287:2254–2257.
- MacHugh DE , Bradley DG . Livestock genetic origins: goats buck the trend. Proc Nat Acad Sci USA. 2001;98:5382–5384.
- Barbana C , Sánchez L , Pérez MD . Bioactivity of α-lactalbumin related to its interaction with fatty acids: a review. Crit Rev Food Sci Nutr. 2011; 51:783–794.
- Ma RN , Deng CJ , Zhang XM , et al. A novel SNP of alpha-lactalbumin gene in Chinese dairy goats. Mol Biol (Mosk). 2010; 44:608–612.
- Qureshi MI , Sabir JSM , Mutawakil MHZ , et al. Review of modern strategies to enhance livestock genetic performance: from molecular markers to next-generation sequencing technologies in goats. J Food Agric Env. 2014;12:752–761.
- Winyoo C , Shannon LK , Jennifer F , et al. Detection of a single nucleotide polymorphism in the human α-lactalbumin gene: implications for human milk proteins. J Nut Biochem. 2005;16:272–278.
- Velliyagounder K , Kaplan JB , Furgang D , et al. One of two human lactoferrin variants exhibits increased antibacterial and transcriptional activation activities and is associated with localized juvenile periodontitis. Infect Immun. 2003;71:6141–6147.
- Ramunno L , Cosenza G , Rando A , et al. The goat as1-casein gene: gene structure and promoter analysis. Gene. 2004;334:105–111.
- Ramunno L , Cosenza G , Rando A , et al. Comparative analysis of gene sequence of goat CSN1S1 F and N alleles and characterization of CSN1S1 transcript variants in mammary gland. Gene. 2005;345:289–299.
- Moatsou G , Hatzinaki A , Samolada M , et al. Major whey proteins in ovine and caprine acid wheys from indigenous greek breeds. Int Dairy J. 2005;15:123–131.
- Prinzenberg EM , Brandt H , Bennewitz J , et al. Allele frequencies for SNPs in the aS1-casein gene (CSN1S1) 5’ flanking region in European cattle and association with economic traits in German Holstein. Livest Prod Sci. 2005;98:155–160.
- Cosenza G , Gallo D , Illario R , et al. A Mval PCR-RFLP detecting a silent allele at the goat alpha-lactalbumin locus. J Dairy Res. 2003;70:355–357.
- Lan XY , Chen H , Tian ZQ , et al. [Correlations between SNP of LALBA gene and economic traits in Inner Mongolian white cashmere goat]. Yi Chuan. 2008;30:69–74. Chinese.
- Falconer DS , Mackay TFC . Introduction to quantitative genetics. 4th ed. Harlow: Pearson; 1996.
- Lan XY , Pan CY , Chen H , et al. An MspI PCR-RFLP detecting a single nucleotide polymorphism at alpha-lactalbumin gene in goat. Czech J Anim Sci. 2007;52:138–142.
- Sun HS , Anderson LL , Yua TP , et al. Neonatal Meishan pigs show POU1F1 genotype effects on plasma GH and PRL concentration. Anim Reprod Sci. 2002;69:223–237.
- Liang Q , Wen D , Xie J , et al. A rapid and effective method for silver staining of PCR products separated in polyacrylamide gels. Electrophoresis. 2014;35:2520–2523.
- Sanger F , Nicklen S , Coulson AD . DNA sequencing with chain terminating inhibitor. Proc Nat Acad Sci USA. 1977;74:5436–5467.
- Zidi A , Casas E , Amills M , et al. Genetic variation at the caprine lactalbumin, alpha (LALBA) gene and its association with milk lactose concentration. Anim Genet. 2014;45:612–613.
- Dettori ML , Pazzola M , Paschino P , et al. Variability of the caprine whey protein genes and their association with milk yield, composition and renneting properties in the Sarda breed. 1. The LALBA gene. J Dairy Res. 2015;82:434–441.
- Zhou JP , Dong CH . Association between a polymorphism of the α-lactalbumin gene and milk production traits in Chinese Holstein cows. Genet Mol Res. 2013;12:3375–3382.
- Dayal S , Bhattacharya TK , Vohra V , et al. Effect of alpha-lactalbumin gene polymorphism on milk production traits in water buffalo. Asian Australas J Anim Sci. 2006;19:305–308.
- El Hanafy AAM , Qureshi MI , Sabir J , et al. Nucleotide sequencing and DNA polymorphism studies of beta-lactoglobulin gene in native Saudi goat breeds in relation to milk yield. Czech J Anim Sci. 2015;60:132–138.