ABSTRACT
A total of 15 fungal isolates were obtained from oil-contaminated sites near the Red Sea in the Yanbu region. Based on the preliminary DCPIP (2,6-dichlorophenolindophenol) assay, three isolates showed promising oil degrading ability. The next-generation sequencing of the ITS-I and ITS-II internal transcribed spacer regions assigned the isolates to Aspergillus and Penicillium. Among these three strains, Y2 (Aspergillus oryzae) was the most efficient, degrading about 99% of the crude oil. The degradation rates were corroborated using spectrophotometric and gas chromatography–mass spectrometry analyses after two weeks of cultivation in Bushnell–Haas medium. All the three strains proved to be potent oil-degrading strains and, hence, can be utilized to degrade oil contaminants.
Introduction
Saudi Arabia, with nearly a quarter of the world's oil reserves, is the largest oil producer and exporter. Oil around the world is transported via pipelines, road, ships and rail, posing great danger to the environment in case of spills. This necessitates the need for developing techniques that are eco-friendly to clean up oil spills. One such method is the use of biological agents due to its efficiency and cost-effectiveness, compared to physico-chemical methods [Citation1].
Crude oil consists of naturally occurring hydrocarbons that are considered environmental pollutants. Its recalcitrance and abundance has been reported in most polluted sites. The degradability of individual hydrocarbon components is influenced by the quality of the hydrocarbon content present in the crude oil. Oil containing large amounts of high-molecular-weight compounds is difficult to degrade biologically because of the complexity of their structure [Citation2]. The chemical composition of oil influences the growth of microbial populations. Microbes that use hydrocarbons as a source of energy thrive under high temperature and salinity conditions [Citation3]. Fungi are known to play a significant role in eliminating hazardous compounds from soil and water contaminated with oil spills, as they inhabit such substrates and utilize hydrocarbons as a source of carbon.
Fungi are known to be one of the best oil-degrading organisms [Citation1,Citation4]. Different studies have identified numerous fungal genera capable of utilizing crude oil as a source of carbon and energy, including Cephalosporium, Rhizopus, Paecilomyces, Alternaria, Mucor, Talaromyces, Gliocladium, Aspergillus, Fusarium, Rhodotolura, Cladosporium, Geotrichum, Penicillium, Torulopsis and Pleurotus [Citation5–7]. The aim of our study was to isolate and identify oil-degrading fungal strains from the coastal area of the Red Sea in Yanbu region contaminated with crude oil, as our previous study showed that this region has potent crude-oil degrading bacterial strains [Citation8].
Materials and methods
Sampling
For the isolation of oil-degrading fungi, eight samples were collected from the shores of the Red Sea, Saudi Arabia. All of the eight collection points were located near the Yanbu industrial Area (24.0833° N: 38.0000° E). Soil samples were taken from 1 to 12 cm below the surface, using a sterile knife.
Isolation and identification
Czapek Dox Agar and potato dextrose agar were used for the isolation and maintenance of fungal isolates. Pure isolates were then tested for their ability to grow on Bushnell–Haas medium, composed of: K2HPO4 (1 g/L), MgSO4 (0.2 g/L), KH2PO4 (1 g/L), CaCl2 (0.02 g/L), NH4NO3 (1 g/L) and FeCl2 (0.05 g/L) with 1% crude oil (Aramco (Yanbu) Company, Saudi Arabia) at 30 °C for one week [Citation9]. Fifteen different isolates were selected for further study based on their ability to grow on Bushnell–Haas medium.
Morphology
Fungal isolates were characterized based on their morphology, e.g. shape, size, spore colour and texture. Fungal mycelia of the isolated strains were observed under light (Olympus BX51, Tokyo, Japan) and scanning electron microscope (Hitachi S 5200, Tokyo, Japan). The microbial growth was determined spectrophotometrically (Jenway 7310, Staffordshire, UK) at 600 nm.
Molecular identification
Approximately 0.5 g of fungal hyphae were taken from each test tube containing fungal isolates. After that, the hyphae were incubated in 100 µL lyticase solution at 30 °C for 60 min. In order to degrade the proteins present in the crude oil samples, 20 µL of Proteinase K (Promega, Madison, WI, USA, 800 µ/mL) was added and incubated at 55 °C for 90 min. The samples were further incubated for two hours at 65 °C. Finally, about 10 µL of each sample was used for polymerase chain reaction (PCR) amplification. The PCR programme was set according to the conditions described by Esteve-Zarzoso et al. [Citation10]. Briefly, for a 25-µL PCR reaction, the primers ITS-I (5′TCCGTAGGTGAACCTTGCGG3′) and ITS-II (5′GCTGCGTTCTTCATCGATGC 3′) were used [Citation11]. The Master Mix (Promega) for PCR contained 0.5 µmol/L primers, 1.5 mmol/L MgCl2, 10 µmol/L deoxynucleoside triphosphates and 1× buffer. The 35 cycles of amplification was carried out in a Thermocycler (Techne, Staffordshire, UK) as follows: 94 °C for 1 min; annealing at 55.5 °C for 2 min and extension at 72 °C for 2 min; and a final extension step at 72 °C for 10 min.
PCR product sequencing and sequence analysis
The results from the sequencing data (Macrogen, Seoul, South Korea) of three isolates were submitted to GenBank (www.ncbi.nlm.nih.gov/genbank/) to obtain the accession numbers (KR137638, KR029081 and KR137639). Previous molecular evolutionary analysis of these strains based on ITS-1 and ITS-II pyrosequencing assigned the strains to Aspergillus niger (KR137638), Aspergillus oryzae (KR029081) and Penicillium commune (KR137639) [Citation12]. The evolutionary distances were computed using the maximum composite likelihood method [Citation13] and were expressed in number of base substitutions per site as a unit of measurement. Evolutionary analysis and phylogenetic tree construction were carried out using MEGA version 4 [Citation14].
2,6-Dichlorophenol indophenol assay
The DCPIP (2,6-dichlorophenol indophenol) technique was used with slight modifications to assess the oil-degrading ability of fungal isolates [Citation15]. After one week of growth, 1 cm2 of fungal hyphae were picked from a Petri dish and transferred to a 500 mL conical flask with 100 mL of Bushnell Haas Media (BHS) containing 0.1% (v/v) of Tween 80, 1% crude oil and 0.6 μg/mL of redox indicator. All the flasks were incubated for two weeks at 30 °C. After two weeks of incubation, the decolourization of DCPIP indicated degradation of crude oil by these isolates and was analysed spectrophotometrically (Jenway 7310) at 420 nm.
Emulsification activity, hydrophobicity and liquid surface tension
To determine the emulsification activity, an equal amount of hexadecane was added to cell-free culture broth, obtained using centrifugation at 10,000 r/min for 5 min. Samples were incubated for 24 hours. The activity was determined as a percentage of the height of the emulsified layer divided by the total height of the liquid column (mm). The surface tension of the degraded sample was analysed using a digital tensiometer. After two weeks of culture, the surface tension was measured by Wilhelmy plate method and was expressed in mN/m units. Fungal adhesion to hydrocarbons (FATH) was assayed according to the method developed by Pruthi and Cameotra [Citation16].
Gravimetric analysis
For gravimetric analysis, an equal volume of dichloromethane was added to the liquid medium containing crude oil. The extracted crude oil was analysed as described in [Citation2].
Gas chromatography–mass spectrometry (GC-MS)
Using GC-MS (Shimadzu GC-Q2010, Tokyo, Japan) flame ionization detector the end product obtained after degradation by Y2 was examined according to the method described by Joo and Kim [Citation17]. After cultivation of isolates in Bushnell–Haas medium at 30 °C at 180 r/min for two weeks, the remaining oil was extracted using a separating funnel in the presence of dichloromethane. Calcium sulphate (drying agent) was used in order to remove the remaining moisture content. The GC-MS programme was set as follows: 60 °C for 2 min, an incremental increase of 6 °C until the temperature reaches 300 °C for 15 min. The operating temperature for the injector was 300 and 320 °C for the detector. Nitrogen was used as a carrier gas. No internal standards were employed. The total time for a GC run was less than 30 min.
Data analysis
Statistical analysis was done using SPSS (version 15).
Results and discussion
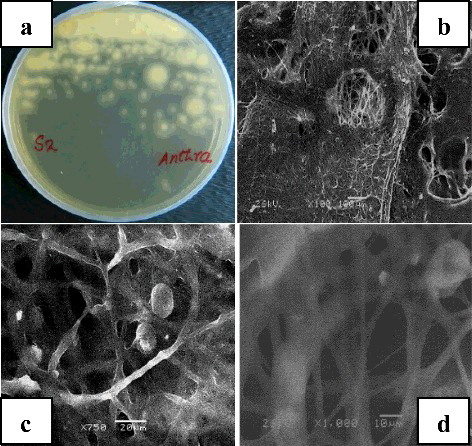
Fungi are known to degrade various kinds of hazardous contaminants. Some species, including Cunninghamella echinulata, Talaromyces spp., Gliocladium spp., Aspergillus spp., Fusarium spp., Rhodotolura spp., Cladosporium spp. and Geotrichum spp., are already known for their ability to degrade oil and other hazardous compounds [Citation18]. In this study, a total of 15 fungal isolates were tested for their ability to biodegrade crude oil, out of which three fungal strains demonstrated perfect biodegradation ability: A. niger (Y1), A. oryzae (Y2) and Penicillium commune (Y4) as shown in . Among these three strains, Aspergillus showed higher degradation ability. shows the morphology of A. oryzae.
Table 1. Effect of hydrocarbons on cell growth, cell surface hydrophobicity, emulsification activity and surface tension after two weeks of incubation at 30 °C.
Figure 1. Morphological characteristics of Aspergillus oryzae (Y2). Fungal isolate from Yanbu soil on nutrient agar plate (a); SEM image of isolate Y2 at 100× magnification (b), 750× magnification (c) and 1000× magnification (d).
Molecular identification
In our previous study [Citation12], the isolates were identified using amplicon pyrosequencing of the ITS-1 and ITS-II internal transcribed spacer regions as A. niger (KR137638), A. oryzae (KR029081) and Penicillium commune (KR137639). The optimal phylogenetic tree of these three strains, with a sum of branch length of 36.19313893, is shown in . The presence of these strains in areas contaminated with crude oil in Yanbu region suggests that they show tolerance to crude oil. Similar studies have also been done recently by different scientists to identify oil-degrading fungi [Citation19–22].
Figure 2. Comparative phylogenetic analysis of the Aspergillus niger (KR137638), Aspergillus oryzae (KR029081) and Penicillium commune (KR137639) [12].
![Figure 2. Comparative phylogenetic analysis of the Aspergillus niger (KR137638), Aspergillus oryzae (KR029081) and Penicillium commune (KR137639) [12].](/cms/asset/5c705afc-605c-49db-a64a-24848606bd4d/tbeq_a_1249407_f0002_oc.jpg)
DCPIP assay
A. oryzae (KR029081) was the most potent of the three isolates, according to the qualitative (DCPIP and spectrophotometry) and quantitative analysis (GC-MS). A similar study on crude-oil-degrading fungi isolated from the Gulf of Mexico was carried out by Al-Nasrawi [Citation23]. There are three main indicators that help identify fungal isolates with the ability to degrade crude oil: decolourization of DCPIP, reduction in the quantity of crude oil and fungal proliferation. The mechanism utilized by fungi to biodegrade crude oil occurs by incorporating an electron acceptor such as DCPIP [Citation23]. DCPIP present in the culture medium serves as an indicator that a degradation process takes place due to the fact that the reagent changes colour from blue (oxidized) to colourless (reduced) [Citation14]. In the presence of A. oryzae, the medium was effectively decolourized, providing evidence that it has a capacity to degrade crude oil.
Cell surface hydrophobicity, emulsification activity and surface tension
In addition to the emulsification activity, FATH and surface tension reduction assays were also performed for the three strains. The data thus obtained from the biosurfactant production assay and ensuing tests are shown in . Isolate A. oryzae (Y2) had higher emulsification activity (E24; 75.6%) and showed a decrease in FATH (26.5%). This suggests production of biosurfactants by this strain and a decrease in the surface tension to a greater extent compared to the other isolates. Our results indicate a direct correlation between cell surface hydrophobicity and biosurfactant production, emulsification activity and crude oil degradation. In a similar study conducted on bacterial isolates from a beach in Brazil, a total of 17 strains were annotated, among which only six were able to reduce the surface tension below 40 mN/m [Citation24]. The production of biosurfactants by microbes is directly proportional to the amount of hydrocarbon degradation. Thus, on the basis of cell-surface hydrophobicity, emulsification activity and surface tension, it was observed and validated that all the three strains were efficient at crude oil degradation.
Spectrophotometric analysis
The spectrophotometric analysis of microbial growth and crude oil biodegradation after cultivation in the presence of 1% crude oil for two weeks showed that the strains Y1 (A. niger), Y2 (A. oryzae) and Y4 (P. commune) exhibited highest level of crude-oil biodegradation ability, degrading 54%, 99% and 48% of the oil, respectively (). Thus, Y2 (A. oryzae), with 99% crude oil degradation, proved to be the most efficient.
A similar study was conducted by Leahy et al. [Citation25], where the hydrocarbon-degrading capabilities of bacteria were documented via spectrophotometry and were analysed at 400 nm wavelength, whereas the degradation kinetics were studied at 540 nm. In another study, A. niger and Penicillium documbens were identified as very efficient fungal isolates, showing high biodegradable activity [Citation25]. A. niger was shown to actively degrade four kinds of hydrocarbons present in oil, Durb oil, Escravos light, Arabian light and Bonny light [Citation26]. Moreover, Aspergillus spp., Penicillium spp., Helminthosporium spp., Rhizopus spp. and Fusarium spp. have also been reported to be efficient metabolizers of hydrocarbons [Citation27,Citation28]. Bartha [Citation29], in their review, mentioned 14 fungal genera isolated from aquatic environments, with the potential to degrade hydrocarbons.
GC-MS analysis
The results from the GC-MS analysis of the metabolic intermediates produced during crude-oil degradation by the most efficient isolate, strain Y2, are shown in and . Major peak compounds were identified at retention time of 9.205, 9.925, 10.595, 11.230, 11.825, 12.388, 12.754 and 12.925 min by comparing the data with standard library compounds such as tridecane, hexadecane, heptadecane, tetracosane, heptacosane, octacosane, nonacosane and hexatriacontane. All the hydrocarbons present in the crude oil that were detected using GC-MS analysis were effectively degraded by the Y2 fungal isolate from Yanbu region.
Figure 3. Bioremediation of crude oil after two weeks of incubation at 30 °C, analysed by GC-MS. (a) without micro-organism (control) and (b) degraded sample using Y2.
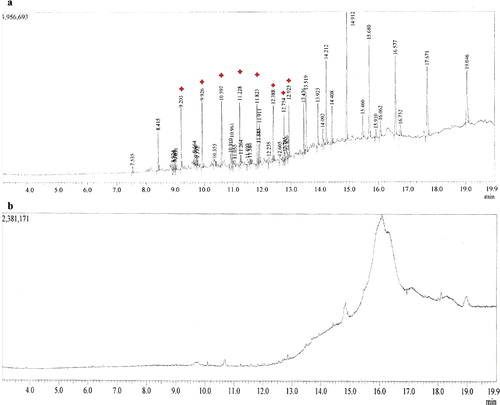
Table 2. Quantitative assessment of crude oil content in the presence of Aspergillus oryzae Y2 along with control.
The results from the spectrophotometric analysis were supported by those obtained by GC-MS analysis, as the isolates efficiently degraded hydrocarbons like tridecane, hexadecane, heptadecane, tetracosane, heptacosane, octacosane, nonacosane, similar to previous reports [Citation30]. For example, Abioye et al. [Citation31] recently reported a Saccharomyces cerevisiae isolate that could give about 49% degradation of crude oil. Previous research at our lab also showed that Yanbu region has oil-degrading micro-organisms that have the ability to degrade nearly 100% of the crude oil in the medium [Citation8,Citation12]. Although different physical and chemical methods are available to remove oil spills, these means of environmental cleaning are not economical. This underlines the importance of isolation and identification of indigenous microbial species that are able to survive harsh environmental conditions and effectively degrade crude oil.
Conclusions
In this study we successfully managed to isolate and identify fungal strains that were able to degrade hydrocarbons present in crude oil. Further study needs to be done to isolate and identify other groups of micro-organisms present in the Yanbu region that are able to utilize long-chain hydrocarbons in the shortest period of time to help expedite the process of bioremediation.
Acknowledgments
The authors are thankful to Aramco (Yanbu) Company for providing the samples. The authors also acknowledge the assistance from the Science & Technology Unit, Deanship of Scientific Research and Deanship of Graduate Studies for the Technical Support, King Abdulaziz University, Jeddah, KSA.
Disclosure statement
The authors declare no conflict of interest.
Additional information
Funding
References
- Ojo OA. Petroleum hydrocarbon utilization by native bacterial population from a wastewater canal in Southwest Nigeria. Afr J Biotech. 2006;5(4):333–337.
- Guo-liang Z, Yue-ting W, Qin M. Biodegradation of crude oil by Pseudomonas aeruginosa in the presence of rhamnolipids. J Zhejiang Uni Sci B. 2005;6(8):725–730.
- Westlake DW, Jobson A, Phillippe R, et al. Biodegradability and crude oil composition. Can J Microbiol. 1974;20:915–928.
- Batelle CD. Mushrooms: higher macrofungi to clean up the environment. Environ Issues. 2000;361–3648.
- Ameen F, Moslem M, Hadi S, et al. Biodegradation of diesel fuel hydrocarbons by mangrove fungi from Red Sea coast of Saudi Arabia. Saudi J Biol Sci. 2016;23(2):211–218.
- Dawoodi V, Mahbobeh M, Arezoo T, et al. The study of heterotrophic and crude oil-utilizing soil fungi in crude oil contaminated regions. J Bioremed Biodeg. [Internet]. 2015 [cited 2016 Apr 17];6:270. Available from: http://www.omicsonline.org/open-access/the-study-of-heterotrophic-and-crude-oilutilizing-soil-fungi-in-crude-oil-contaminated-regions-2155-6199.1000270.php?aid=40759.
- Zhang JH, Quan-Hong X, Hui G, et al. Degradation of crude oil by fungal enzyme preparations from Aspergillus spp. for potential use in enhanced oil recovery. J Chem Technol Biotechnol. 2016;91:865–875.
- EL-Hanafy AA, Anwar Y, Mohamed SA, et al. Isolation and identification of bacterial consortia responsible for degrading oil spills from the Coastal area of Yanbu, Saudi Arabia. Biotechnol Biotechnol Equip. 2015;30(1):69–74.
- Anupama M, Padma S. Studies on biodegradaction of crude oil by Aspergillus niger. South Pacific J Nat Sci. 2009;27:57–60.
- Esteve-Zarzoso B, Belloch C, Uruburu F, et al. Identification of yeasts by RFLP analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers. Int J Syst Bacter. 1999;49:329–337.
- White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York (NY): Academic Press; 1990. p. 315–322.
- EL-Hanafy AA, Anwar Y, Mohamed SA, et al. Isolation and molecular identification of two fungal strains capable of degrading hydrocarbon contaminants on Saudi Arabian environment. Int J Biol Biomolecular Agri Food Biotech Engineer. 2015;9:1169–1172.
- Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA. 2004;101:11030–11035.
- Tamura K, Dudley J, Nei M, et al. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599.
- Hanson KG, Desai JD, Desai AJ. A rapid and simple screening technique for potential crude oil degrading microorganisms. Biotechnol Tech. 1993;7:745–748.
- Pruthi V, Cameotra SS. Rapid identification of biosurfactant-producing bacterial strains using a cell surface hydrophobicity technique. Biotechnol Tech. 1997;11:671–674.
- Joo MH, Kim JY. Characteristics of crude oil biodegradation by biosurfactant-producing bacterium Bacillus subtilis JK-1. J. Korean Soc Appl Biol Chem. 2013;56:193–200.
- Cutright TJ, Lee SG. In-situ soil remediation-bacteria or fungi. Ener Sources. 1995;17:413–419.
- Hu HL, van den BJ, Gruben BS, et al. Improved enzyme production by co-cultivation of Aspergillus niger and Aspergillus oryzae and with other fungi. Int Biodeterior Biodegrad. 2011;65:248–252.
- Ihsan FHA. Ability of some fungi isolated from a sediment of Suq-Al Shuyukh marshes on biodegradation of crude oil. Int J Curr Microbiol App Sci. 2015;4(1):19–32.
- Maddela NR, Masabanda M, Leiva-Mora M. Novel diesel-oil-degrading bacteria and fungi from the Ecuadorian Amazon rainforest. Water Sci Technol. 2015;71:1554–1561.
- Maddela NR, Scalvenzi L, Perez M, et al. Efficiency of indigenous filamentous fungi for biodegradation of petroleum hydrocarbons in medium and soil: laboratory study from Ecuador. B Environ Contam Tox. 2015;95:385–394.
- Al-Nasrawi H. Biodegradation of crude oil by fungi isolated from Gulf of Mexico. J Bioremed Biodegrad. [Internet]. 2012 [ cited 2016 Apr 17];3:147. Available from: http://www.omicsonline.org/biodegradation-of-crude-oil-by-fungi-isolated-from-gulf-of-mexico-2155-6199.1000147.php?aid=5829.
- Batista SB, Mounteer A, Amorim FR, et al. Isolation and characterization of biosurfactant/bioemulsifier-producing bacteria from petroleum contaminated sites. Bioresour Technol. 2006;97:868–875.
- Leahy JG, Tracy KD, Eley MH. Degradation of mixtures of aromatic and choroaliphatic hydrocarbons by aromatic hydrocarbon-degrading bacteria. FEMS Microbiol Ecol. 2003;43:271–276.
- Gesinde AF, Agbo EB, Agho MO, et al. Bioremediation of some Nigerian and Arabian crude oils by fungal isolates. Int J Pure Appl Sci. 2008;2:37–44.
- Obire O, Anyanwu EC. Impact of various concentrations of crude oil on fungal populations of soil. Int J Environ Sci Technol. 2009;6:211–218.
- Adekunle AA, Oluyode TF. Biodegradation of crude petroleum and petroleum products by fungi isolated from two oil seeds (melon and soybean). J Environ Biol. 2005;26(1):37–42.
- Bartha R. The microbiology of aquatic oil spills. Adv Appl Microbiol. 1977;22:225–266.
- Norman RS, Moeller P, McDonald TJ, et al. Effect of pyocyanin on a crude-oil-degrading microbial community. Appl Environ Microbiol. 2004;70(7):4004–4011.
- Abioye OP, Akinsola RO, Aransiola SA, et al. Biodegradation of crude oil by Saccharomyces cerevisiae isolated from fermented zobo (locally fermented beverage in Nigeria). Pak J Biol Sci. 2013;15:2058–2061.
