ABSTRACT
The success of the breeding programmes depends on the dimension of genetic variability. Simple sequence repeats (SSR) have been extensively used in the studies including genetic characterization of many plant species. The objective of this study was to investigate the genetic variation in 22 faba bean genotypes, 18 of which were originated from International Center for Agricultural Research in the Dry Areas (ICARDA) and 4 of which were cultivated genotypes in Turkey, using SSR markers. Of 41 SSR markers used, 25 produced bands. As a result of SSR amplification, a total of 39 bands, 25 of which were polymorphic and 14 of which were monomorphic, were obtained. The mean gene diversity and polymorphism information content values were 0.27 and 0.24, respectively. The faba bean genotypes cultivated in Turkey had greater genetic diversity than those that originated from ICARDA. The faba bean genotypes FLIP10- and FLIP03- were successfully separated, using the un-weighted pair group method with arithmetic average dendrogram constructed via Jaccard similarity coefficients. These results were further supported by factor analysis substantially. The results indicated that there is sufficient genetic diversity among the tested faba bean genotypes (especially cultivated in Turkey) and could be used in faba bean breeding programmes.
KEYWORDS:
Introduction
Faba bean (Vicia faba L.) was one of the oldest cultivated crops in the Near East region and has been grown for both its fresh pods and dry seeds. Its high protein content (mean 25%) makes faba bean a valuable food for human and animal nutrition [Citation1]. The leading faba bean producer countries are China, Etiyopya, France and Egypt. On the other hand, in Turkey it occupies third rank in food legume, following chickpea, lentil and common bean [Citation2]. Faba bean does not take well-deserved place due to the consumer habit. Actually, the number of registered varieties has been very limited depending on the insufficient faba bean breeding programmes.
One of the two fundamental rules in plant breeding is to create or to exploit genetic variation and the other is selection of appropriate plants. The most efficient way to overcome the biotic and abiotic limitations to faba bean production is to develop the varieties suitable to the environments in which faba bean is cultivated. It is undoubted that the genetic variation/diversity made by introduction, hybridization or mutation must be revealed to select the genotypes resistant to both pests and disease or adapted to environmental conditions such as temperature and soil.
A number of various markers have been used for revealing the genetic diversity in crop plants. Despite being used for a long time [Citation3], the morphological and biological markers are sensitive to the environmental conditions and have some limitations [Citation4]. That is why various DNA markers such as random amplified polymorphic DNA, restriction fragment length polymorphism, amplified fragment length polymorphism (AFLP) and simple sequence repeats (SSR) have been developed. Molecular markers are rapid, are not influenced from environmental conditions and are secure for selecting the important agricultural characteristics. Therefore, they have been used in faba bean breeding to detect genetic variation or similarities [Citation5–7].
SSR markers or microsatellites have been used in many crops since they are highly polymorphic, polymerase chain reaction (PCR) based and easily transferable [Citation8]. These markers have been successfully used in faba bean to build gene map [Citation9], to assess resistance to Orobanche crenata and to determine genetic diversity [Citation10,Citation11]. Furthermore, determination of genetic similarity and phylogenetic relationships among Vicia species and varieties has been successfully accomplished.
This study was carried out to determine the genetic profile of 22 faba bean genotypes, 18 of which were originated from International Center for Agricultural Research in the Dry Areas (ICARDA) and 4 of which were cultivated genotypes in Turkey based on SSR markers in order to measure the extent of genotypic differences, genetic relationship and to assist in broadening the germplasm base of future faba breeding programmes.
Materials and methods
Plant material
A total of 22 faba bean genotypes, 18 of which were originated from ICARDA and 4 of which from Turkey, were used in this study. The four faba bean genotypes originated from Turkey comprised of two nationally registered cultivars (Kıtık-2003 and Eresen-87) and two local landraces (Antakya and Yayladağı). The names, pedigree and origin of the faba bean genotypes used in this study are presented in
Table 1. The names, pedigree and origin of faba bean genotypes.
DNA extraction
A bulk of 2-week-old leaves from each faba bean genotype were employed for extracting DNA as reported by Dellaporta et al. [Citation12] and Doyle [Citation13] with minor modifications. Genomic DNA was quantified on nanodrop (ACTGene UVS-99, USA), at 260/280 nm. The quality of extracted DNA was confirmed on 0.8% agarose gel electrophoresis.
SSR primers and SSR PCR amplification
The sequences of the 25 SSR primers are listed in . PCR analysis was performed in a final volume of 20 µL containing 30 ng of template DNA, 2.5 mmol/L MgCl2, 2 µmol/L primer, 1 unit of Taq polymerase and 2 mmol/L deoxynucleotide triphosphates (dNTP) in 10× reaction buffer. DNA amplifications were accomplished by the Multi Gene Thermal Cycler (Labnet International, USA), and commenced with denaturation for 5 min at 95 °C, followed by 35 cycles of 30 s denaturation at 94 °C and 30 s annealing at 50–60 °C, 1 min extension at 72 °C, and final extension for 10 min at 72 °C. After amplification, the reaction products were electrophoresed on 3% Nu Micropor agarose (Prona, Spain) agarose gels containing 1× Tris-borate-Ethylenediaminetetraacetic acid (EDTA) buffer. The genomic DNA was stained with 1 µg/mL ethidium bromide. The gels were run at 140 V for 80 min. Gel photos were taken under UV light using the DNR MiniLumi (DNR Bio-Imaging Systems, Israel) gel documentation system.
Table 2. The name and sequences of 25 SSR primers used in the study.
Statistical analysis
Data scores were obtained according to the presence/absence criterion, which regarded presence of band as ‘1’ and absence of band as ‘0.’ The pair-wise similarity matrix was built using Jaccard coefficient calculated according to the formula Jij = a/a + b + c, where Jij is the similarity between two individuals i and j, a is the number of bands present in the both individuals, b is the number of bands observed only in the individual i and c is the number of bands observed only in the individual j. Dendrogram was constructed by the un-weighted pair group method with arithmetic average (UPGMA) method [Citation14] using NTSYSpc v2.1 software. Polymorphism information content (PIC) for each primer was calculated according to the formula [Citation15], where PIC is polymorphism information content, pi and pj represent frequency of i. and j. Alleles, respectively. Both the PIC and Nei's (1973) gene diversity of the primers were calculated by the program Powermarker v3.25 [Citation16]. Nei's (1973) gene diversity and Shannon index [Citation17] values of the faba bean genotypes were calculated by using Popgen v1.32 [Citation18] program. Exploratory factor analysis (FA) was performed on SSR data by Statistica v10 statistic program. The principal axis method was used to extract the factors followed by a varimax rotation. Only the factors whose eigenvalues were greater than 1 were retained for rotation.
Results and discussion
Assessment of genetic diversity and revealing of the genetic relationships in the faba bean germplasm are required to exploit these genetic resources in crop breeding programmes. Because of the difficulties to achieve interspecific crosses or to produce transgenic lines with V. faba, only natural variability or mutation is available to breeders [Citation19]. Taking increasing number of faba bean genotypes into account, discrimination of them by only morphological characters is troubled due to the fact that these characters can be affected by environmental conditions [Citation7]. Therefore, numerous studies based on molecular markers have been conducted worldwide to reveal genetic relatedness in faba bean. On the other hand, according to our knowledge, there have not been any accessible studies concerning DNA markers except for protein marker in faba bean in Turkey [Citation20]. SSR markers or microsatellites have higher efficiency for within-population diversity and similar efficiency for among-population diversity in comparison to dominant markers [Citation21]. Furthermore, they are powerful tool to determine genetic diversity and polymorphism [Citation22,Citation23]. Therefore, we analysed genetic diversity and population structure among 22 faba bean genotypes by using 25 SSR markers, so that they can facilitate selection of different parents for future crossing programmes.
SSR marker system was successfully used to determine genetic diversity among ICARDA and Turkish faba bean germplasm. Forty-one SSR primers were screened in a preliminary study. Sixteen of them did not yield clear bands; therefore, the remaining 25 primers () were employed in this study. Taking into account that the primers DMA093, M46, DMA042, DMA101, DMA107, M10, DMA099, M17, DMA043 and M14 did not produce polymorphic bands, the mean value of Nei's (1973) gene diversity (h), PIC value, the number of polymorphic bands (npb), the number of monomorphic bands (nmb) and polymorphism (%) were calculated by excluding these primers. Total number of allele was 39 and the number of allele per primer or locus was 1.6 ranging from 1 to 3 depending on the SSR primers across the 22 genotypes studied. A sample amplification pattern of the primer DMA045 is shown in
Figure 1. A sample SSR amplification pattern of the primer DMA045 across faba bean genotypes. The numbers show the faba bean genotypes listed in .
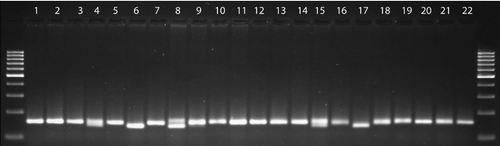
Gene diversity or expected heterozygosity often indicates genetic diversity in a population [Citation24]. The Nei's (1973) gene diversity (h) showed a significant level of variation. The mean Nei's (1973) gene diversity (h) of the loci producing polymorphic bands in the 22 faba bean genotypes was 0.42. The primers DMA030 and DMA070 showed the highest gene diversity with the values of 0.70 and 0.66, respectively, which was supported by high number allele (). On the other hand, the remaining primers showed lower diversity. Ovesna et al. [Citation25] also reported that high number of allele was in accordance with high gene diversity in the study aiming to identify of Czech garlic varieties using 14 SSR markers. The number of allele and gene diversity in the present study coincided with Ma et al. [Citation26], who found the former between 2 and 9, and the latter between 0.0476 and 0.8304 in the study employing 32 accessions of V. faba and 21 microsatellite markers. Moreover, Gong et al. [Citation27] stated the number of allele varying from 1 to 3, and expected heterozygosity from 0 to 0.64 in the study comprising 29 faba bean genotypes and 11 microsatellite markers, which was very similar to ours.
Table 3. Nei's (1973) gene diversity (h), PIC value, the number of polymorphic bands (npb), number of monomorphic bands (nmb), the number of allele (na) and polymorphism (%) obtained from amplification of the SSR primers across 22 faba bean genotypes.
The PIC value can be used to estimate usefulness of SSRs for identifying cultivars [Citation28]. The PIC value per primer varied from 0.00 to 0.64 with an average of 0.37. The 10 primers had a PIC value of zero due to producing only one monomorphic band. The primers DMA030, DMA070, DMA044 and M20 showed the highest PIC values in this study (). Akash and Myers [Citation29], who developed faba bean expressed sequence tag (EST)-SSR markers using 13 V. faba and seven Vicia species to study their transferability among seven related Vicia spp., also found the PIC values of the primers DMA030 and DMA070 to be high (0.72 and 0.69, respectively). However, they reported the lower PIC values for the primer DMA044 with a value of 0.20 in comparison to our study. Gong et al. [Citation11] reported PIC values varied from 0.0644 to 0.4278 with an average of 0.2919 in their study in order to investigate the genetic variation of 29 faba bean cultivars from China and Europe using 11 EST-SSR markers. This mean PIC value was similar to ours. Zeid et al. [Citation10] found PIC values ranging from 0.16 to 0.72, which extensively overlapped our results. Taking the gene diversity, the PIC value and the number of alleles into consideration, the most two informative markers were DMA030 and DMA070. The polymorphism (%) of the primers varied from 0% to 100% with an average of 87.8%, which indicated high variability in faba bean genotypes.
The effective number of alleles (ne), Nei's (1973) gene diversity (h) and the Shannon Index (I) varied depending on the genotypes of faba bean. The genotypes Eresen-87, FLIP03-38FB and Yayladağı showed the highest genetic diversity with regard to these parameters across the faba bean genotypes (). Indeed, this was not surprising since the breeding lines from ICARDA were selected for resistance or tolerance to chocolate spot diseases. In addition, the ICARDA derived lines named with Flip10 and Flip03 prefixes were explicitly separated in this respect. The mean Nei's (1973) gene diversity (0.40) in the faba bean genotypes was similar to that obtained by Yahia et al. [Citation30], who reported a mean value of 0.43, using 16 Tunisian faba bean local populations and 16 polymorphic SSR markers. In the present study, mean effective number of alleles (1.67) was lower than that (2.25) obtained by Yahia et al. [Citation30] and relatively similar to Wang et al. [Citation31], who studied genetic diversity and relatedness of 802 faba bean genotypes using 11 inter-simple sequence repeat (ISSR) primers and found a mean value of 1.3888. This might be due to different number and origin of faba bean genotypes used in the three studies. Compared to the study of Zong et al. [Citation6], which investigated molecular variations of 243 faba bean germplasm originated from different geographical areas using 10 AFLP primer pairs, results of this study also revealed the pattern of genetic variation across the faba bean genotypes. They obtained lower range of Nei's (1973) gene diversity (0.0433–0.1724) and Shannon Index (0.0639–0.2535) than ours, which might be explained by the different DNA markers used. Ouji et al. [Citation32] investigated molecular diversity of nine faba bean populations using seven isozyme systems and found that Nei's (1973) gene diversity was 0.204. Among the 22 genotypes, the ones originated from Turkey exhibited the highest level of variability in terms of Nei's (1973) gene diversity (h) and Shannon Index (I) with the mean values of 0.44 and 0.63, respectively, whereas the genotypes named with Flip10 prefix showed the lowest level of variability ().
Table 4. Number of alleles (na), effective number of alleles (ne), Nei's (1973) gene diversity (h) and Shannon index (I) values of faba bean genotypes.
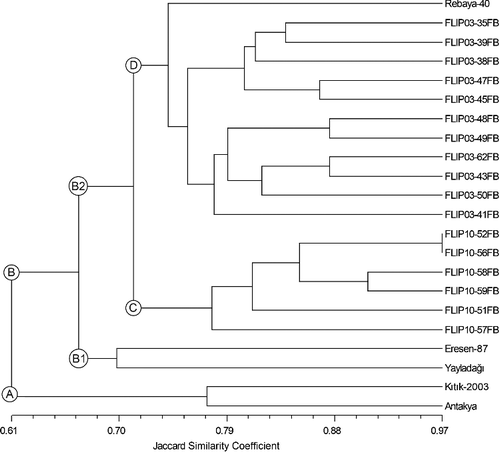
The similarity matrix, estimated from Jaccard coefficients, was used for building dendrogram by UPGMA method using 22 faba bean genotypes. The dendrogram generally agreed with the pedigree and origin of these genotypes (). The faba bean genotypes were grouped into two main clusters, namely A and B in the dendrogram. The cluster A was comprised of the two faba bean genotypes, namely Kıtık-2003 and Antakya, while the cluster B contained all of the remaining genotypes. Particularly the breeding lines derived from same crossing programme of ICARDA were gathered in the same group. It is noted that the genotypes named with Flip10 and Flip03 prefixes were successfully separated in the clusters C and D, respectively. However, the variety Rebaya-40, originated from Egypt, formed a distinct group in the cluster D and was loosely grouped with the FLIP03 prefixed lines probably due to its different origin, namely Egypt. It was also revealed by the dendrogram that all the genotypes originated from Turkey were not grouped in the same cluster while the genotype Antakya was grouped with the genotype Kıtık-2003 and Yayladağı with Eresen-87 (). This was probably because pedigrees of the genotypes Eresen-87 and Kıtık-2003 were not common despite having been developed in different years in the Aegean Agricultural Research Institute, Turkey. Moreover, the fact that the genotypes Antakya and Yayladağı were local populations with unknown pedigree may cause that they were separately clustered in the dendrogram. The least genetic similarity (0.47) occurred between the genotypes Antakya and FLIP03-38FB, whereas the greatest one (0.97) occurred between FLIP10-56FB and FLIP10-52FB (similarity table is not given).
Figure 2. UPGMA dendrogram of 22 faba bean genotypes based on Jaccard similarity coefficients converted from SSR marker data.
Exploratory FA is thought to exert causal influence on observed variables and reveal the underlying construct (the factor) which produces scores on the variables [Citation33]. Therefore, the FA was performed for SSR data to reduce the variables which accounted for most of the variance. The FA extracted four factors eigenvalue of which was greater than 1. The first four factors accounted for 67.43% of the total variation for SSR analysis (). As noted in , all the faba bean genotypes, except for very few ones, were separately and considerably placed in the four factors by the factor loadings greater than 0.45. In case of FLIP03 prefixed lines apart from FLIP03-49FB, they had greater loading on factor1 and were grouped together. On the other hand, factor2 separated FLIP10 prefixed lines except for FLIP10-57FB from the others. In case of Turkey originated genotypes apart from Eresen-87, factor3 distinguished them successfully. Finally, the variety Rebaya-40 had a greatest loading on factor4. Ordination methods have been frequently used to support cluster analysis in some researches. Some of them in general agreed with cluster analysis [Citation7,Citation31,Citation34] although the others supported cluster analysis moderately [Citation11,Citation35]. In the case of this study, the exploratory FA extensively corresponded well with the cluster analysis in grouping the faba bean genotypes. As shown in , the genotypes factor loading which was greater than 0.45 in a factor was similarly clustered also in the dendrogram. However, the variety Eresen-87 was grouped with the FLIP03 prefixed genotypes in the factor1 in FA despite clustered with the genotype Yayladağı in the dendrogram.
Table 5. Both individual and cumulative eigenvalues and variance explained by the first four factors of SSR data in faba bean genotypes.
Table 6. Factor loadings (varimax) of 22 faba bean genotypes on the first four factors by SSR data.
Conclusions
Only a few faba bean cultivars were registered in Turkey. However, both consumer and farmer preferences encourage the faba bean breeders to breed more attractive faba bean cultivars. Therefore, it is important to know the genetic diversity in faba bean genotypes to develop the new faba bean cultivars with both high yield and quality. The results of the current study showed good enough genetic diversity in faba bean genotypes, particularly the ones cultivated in Turkey, and could be facilitated in the planning of future faba bean breeding programmes.
Acknowledgments
We appreciate Mr. Emre İLHAN for his technical assistance in laboratory and the University of Mustafa Kemal for financial support.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Musallam IW, Al-Karaki GN, Ereifej KI. Chemical composition of faba bean genotypes under rainfed and irrigation conditions. Int J Agric Biol. 2004;6:359–362.
- Fao. Faostat. 2012. Available from: http://faostat.fao.org/site/567/DesktopDefault.aspx? PageID=567 (accessed 18 May 2015).
- Ferguson ME, Robertson LD. Morphological and phenological variation in the wild relatives of lentil. Genet Resour Crop Evol. 1999;46:3–12.
- Lee M. The phenotypic and genotypic eras of plant breeding. In: Lamkey KR, Lee M, editors. Plant breeding: the Arnel R. Hallauer international symposium. Ames (IA): Blackwell Publishing Ltd.; 2006. p. 213–218.
- Gresta F, Avola G, Albertini E, et al. A study of variability in the Sicilian faba bean landrace ‘Larga di Leonforte’. Genet Resour Crop Evol. 2010;57:523–531.
- Zong X, Liu X, Guan J, et al. Molecular variation among Chinese and global winter faba bean germplasm. Theor Appl Genet. 2009;118:971–978.
- Terzopoulos PJ, Bebeli PJ. Genetic diversity analysis of Mediterranean faba bean (Vicia faba L.) with ISSR markers. Field Crop Res. 2008;108:39–44.
- Lichtenzveig J, Scheuring C, Dodge J, et al. Construction of BAC and BIBAC libraries and their applications for generation of SSR markers for genome analysis of chickpea, Cicer arietinum L. Theor Appl Genet. 2005;110:492–510.
- Pozarkova D, Koblizkova A, Roman B, et al. Development and characterization of microsatellite markers from chromosome 1-specific DNA libraries of Vicia faba. Biol Plant. 2002;45:337–345.
- Zeid M, Mitchell S, Link W, et al. Simple sequence repeats (SSRs) in faba bean: new loci from Orobanche-resistant cultivar 'Giza 402'. Plant Breeding. 2009;doi:10.1111/j.1439-0523.2008.01584.x
- Gong Y, Xu S, Mao W, et al. Genetic diversity analysis of faba bean ( Vicia faba L.) based on EST-SSR markers. Agric Sci China. 2011;10:838–844.
- Dellaporta S, Wood J, Hicks J. A plant DNA mini-preparation: version II. Plant Mol Biol Rep. 1983;1:19–21.
- Doyle JJ. Isolation of plant DNA from fresh tissue. Focus (San Francisco, Calif.) 1990;12:13–14.
- Sneath PHA, Sokal RR. Numerical taxonomy: the principle and practice of numerical classification. San Francisco: W.F. Freeman & CO; 1973.
- Botstein D, White RL, Skolnick M, et al. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980;32:314–331.
- Liu K, Muse SV. PowerMarker: integrated analysis environment for genetic marker data. Bioinformatics. 2005;21:2128–2129.
- Lewontin RC. The apportionment of human diversity. Evol Biol. 1972;6:381–398.
- Yeh FC, Boyle TJB. Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg J Bot. 1997;129:157–163.
- Duc G, Bao S, Baum M, et al. Diversity maintenance and use of Vicia faba L. genetic resources. Field Crop Res. 2010;115:270–278.
- Binici B. Determination of genetic similarity by seed storage protein electrophoresis in some fava bean (Vicia faba L.) cultivar and lines and quality criteria [dissertation]. Hatay: University of Mustafa Kemal; 2013.
- Nybom H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol. 2004;13:1143–1155.
- Sethy NK, Choudhary S, Shokeen B, et al. Identification of microsatellite markers from Cicer reticulatum: molecular variation and phylogenetic analysis. Theor Appl Genet. 2006;112:347–357.
- Serret MD, Udupa SM, Weigand F. Assessment of genetic diversity of cultivated chickpea using microsatellite-derived RFLP markers: implications for origin. Plant Breed. 1997;116:573–578.
- Nassiry MR, Javanmard A, Tohidi R. Application of statistical procedures for analysis of genetic diversity in domestic animal populations. Am J Anim Vet Sci. 2009;4:136–141.
- Ovesna J, Leisova-Svobodova L, Kucera L. Microsatellite analysis indicates the specific genetic basis of Czech bolting garlic. Czech J Genet Plant Breed. 2014;50:226–234.
- Ma Y, Yang T, Guan J, et al. Development and characterization of 21 EST-derived microsatellite markers in Vicia faba (fava bean). Am J Bot. 2011;98:e22–e24.
- Gong Y, Xu S, Mao W, et al. Generation and characterization of 11 novel EST-derived microsatellites from Vicia faba (Fabaceae). Am J Bot. 2010;97:e69–e71.
- Martin JP, Borrego J, Cabello F, et al. Characterization of Spanish grapevine cultivar diversity using sequence-tagged microsatellite site markers. Genome. 2003;46:10–18.
- Akash MW, Myers GO. The development of faba bean expressed sequence tag–simple sequence repeats (EST-SSRs) and their validity in diversity analysis. Plant Breed. 2012;131(4):522–530.
- Yahia Y, Hannachi H, Monforte AJ, et al. Genetic diversity in Vicia faba L. populations cultivated in Tunisia revealed by simple sequence repeat analysis. Plant Genet Resour. 2014;12:278–285.
- Wang HF, Zong XX, Guan JJ, et al. Genetic diversity and relationship of global faba bean (Vicia faba L.) germplasm revealed by ISSR markers. Theor Appl Genet. 2012;124: 789–797.
- Ouji A, Suso MJ, Rouaissi M, et al. Genetic diversity of nine faba bean (Vicia faba L.) populations revealed by isozyme markers. Genes Genom. 2011;33:31–38.
- Tabachnick BG, Fidell LS. Using multivariate statistics. 6th ed. Boston (MA): Pearson; 2013.
- Tantasawat P, Trongchuen J, Prajongjai T, et al. Variety identification and comparative analysis of genetic diversity in yardlong bean (Vigna unguiculata spp. sesquipedalis) using morphological characters, SSR and ISSR analysis. Sci Hortic. 2010;124:204–216.
- Coulibaly S, Pasquet RS, Papa R, et al. AFLP analysis of the phenetic organization and genetic diversity of Vigna unguiculata L. Walp. reveals extensive gene flow between wild and domesticated types. Theor Appl Genet. 2002;104:358–366.
