ABSTRACT
Eighteen Bulgarian newly bred grapevine varieties obtained by interspecific crossing were analysed through microsatellite markers in order to determine their genetic identity as well as the presence of resistance-related alleles linked to Rpv3, Rpv10 and Rpv12 loci. The levels of resistance of the investigated cultivars to downy mildew were assessed by leaf disk assay and were scored according to OIV descriptor 452. Seven isolates of Plasmopara viticola collected in different regions of Bulgaria were characterized and discriminated with seven microsatellite markers and were used in the assays. Analysis of variance showed significant differences in resistance to downy mildew among the analysed varieties. A clear correlation between the presence of particular Rpvs and the level of resistance was determined. Five cultivars, ‘Droujba’, ‘Garant’ and ‘Plevenska rosa’ harbouring Rpv3 and Rpv12, ‘Kajlashki rubin’ harbouring Rpv12 and ‘Slava’, Rpv10 and Rpv3, showed high level of resistance with mean OIV452 values ranging from 6.5 to 8.25 over all seven P. viticola isolates. The level of resistance of the remaining 14 cultivars, which carry only Rpv3 inherited from cultivar Villard Blanc, was found to vary significantly among the isolates with mean OIV452 values between 1.75 and 4.8 over all used P. viticola isolates. The results obtained in this study favoured five genotypes remarkable for their high resistance to downy mildew and very good quality of grape and wine. These varieties represent a valuable material for pyramiding of resistance through marker-assisted selection.
Introduction
Downy mildew, caused by the Oomycete Plasmopara viticola (Berk. and Curt.) Berl. and de Toni., is one of the most severe diseases in grapevine. It causes defoliation of grapes, damage of berries and even crop loss, thus leading to huge injury to viticulture. Currently, the disease is controlled by treatment with fungicides, which, in favourable conditions, can reach up to 14 treatments per year in Bulgaria. The development of varieties with increased tolerance to downy mildew is an alternative way of controlling the disease and decreasing fungicide application. Since the European vine Vitis vinifera is sensitive to P. viticola, other resistant Vitis species are used for introduction of such resistance in V. vinifera. Presently, a few sources of resistance to P. viticola in grapevine are known such as North American and Asian Vitis species. In the last two decades, owing to the development of DNA marker systems and the sequencing of the grapevine genome, a number of genetic determinants of resistance to P. viticola, named Rpv (resistance to Plasmopara viticola) loci, have been discovered in these species. Information about 15 Rpv loci underlying resistance to P. viticola is available in the Vitis International Variety Catalogue (VIVC) [Citation1]. Molecular markers associated with these Rpv loci can be used in breeding programmes for marker-assisted selection (MAS) as well as for evaluation of grapevine genetic resources for the presence of such loci.
The intensive breeding for resistance to P. viticola, which started after the invasion of P. viticola in Europe in the second half of the nineteenth century, exploited exclusively North American Vitis species as a source of resistance. This resulted in development of a large number of breeding lines sharing a few Rpv3 haplotypes, transmitted from these species, leading to the risk of appearance of new strains of P. viticola that can overcome the resistance determined by this locus [Citation2]. Indeed, Peressotti et al. [Citation3] proved that a particular strain of P. viticola evades the resistance in cultivar ‘Bianca’, which harbours the most spread Rpv3 haplotype. For this reason, modern breeding aims at development of breeding lines/cultivars with durable resistance by combining two or three resistant loci of different origin in one genotype [Citation4–6].
Breeding for resistance to P. viticola in Bulgaria started in the second half of the twentieth century, when Seyve Villard 12-375, a French hybrid with North American Vitis species ancestry, also known as Villard Blanc, was used as a source of resistance to downy mildew. Later on, V. amurensis or its descendant cultivars, ‘Zarya Severa’ and’ Tsvetochnyi’, were also included in the development of newly bred varieties. The resistance of these hybrids to downy mildew was evaluated in the field under the conditions of natural P. viticola infection. The leaf disk assay allows more reliable assessment of the resistance to downy mildew, because the test is performed in controlled conditions, i.e. temperature, humidity, light and concentration of the P. viticola sporangia in the suspension used for inoculation of the leaf discs, thus avoiding the influence of climatic conditions.
Currently, the development of P. viticola simple sequence repeat (SSR) markers by Delmotte et al. [Citation7] and Rouxel et al. [Citation8] allows the determination of genetic diversity and discrimination between different isolates of P. viticola, which can be used in the leaf disk assay.
In this study, we characterized 18 newly bred Bulgarian varieties originating from interspecific crossings in regard to their genetic identity, level of resistance to downy mildew and presence of Rpv loci. The level of resistance of the investigated varieties to seven P. viticola isolates collected from different regions of Bulgaria was assessed by the leaf disk assay. SSR markers were applied for: (1) determination of the genetic authenticity of the analysed varieties, (2) detection of resistance-related alleles at SSR markers linked to three Rpv loci in the investigated genotypes and (3) characterization of and discrimination between P. viticola isolates.
Materials and methods
Plant material
The investigated eighteen newly bred varieties are grown in the grapevine collection of the Institute of Viticulture and Enology, Bulgaria. They were obtained by interspecific crossing and include SV12375 or SV12375 and ‘Zarya Severa’, or SV12375 and ‘Tsvetochnyi’ or V. amurensis Ruprecht in their pedigree (Table S1 in the Online Supplementary Appendix) [Citation1,9–12]. The names of the cultivars and the source of resistance are shown in . Eight biological replicates per variety were propagated from cuttings and were planted in greenhouse. The cultivars ‘Bianca’, ‘Kunbarat’ and V. amurensis Ruprecht were used as controls for the presence of Rpv3, Rpv12 and Rpv10, respectively.
Table 1. List of 18 newly bred varieties, their names, cultivar number according to VIVC, source of resistance according to their pedigree and allele size (bp) at markers associated with Rpv3, Rpv10 and Rpv12 loci.
Plasmopara viticola isolates
Infected grapevine leaves at the oil spot stage were collected from the field in seven different locations in Bulgaria and the obtained P .viticola isolates were numbered from 1 to 7 accordingly: Galabnik (1), Dobromirka (2), Pleven (3), Pomorie (4), Ruse (5), Sofia (6) and Vidin (7). After incubation of the infected leaves at 22 °C, the obtained sporangia were propagated by infection of detached leaves of sensitive V. vinifera varieties for further use in leaf disk assay. Part of the infected leaves was stored at −20 °C.
Phenotypic analysis
The level of resistance of the investigated cultivars was analysed by leaf disk assay. The fourth and fifth leaves from the shoot apex were detached. Four leaf disks of 18-mm diameter were excised with a cork borer and were placed with the abaxial surface up on 0.8% water agar in Petri dishes. The disks were artificially inoculated with 40 µL drops of sporangia suspension with a concentration of 20,000 sporangia per millilitre. The Petri dishes were incubated in a growth chamber for 5–7 days at 22 °C with a photoperiod of 16 h/8 h and high humidity. Evaluation of the degree of resistance was performed according to Schwander at al. [Citation6]. The intensity of sporangiophores was assessed at 5–7 days by categorical values from 1 to 9 (9: no, 7: one to five, 5: six to twenty, 3: more than twenty, 1: dense sporangiophore carpet) and classified according to the OIV (Organisation Internationale de la Vigne et du Vin) descriptor 452 [Citation13]. Two assays per each isolate were performed. The resistant cultivar ‘Kunbarat’ and the cultivar ‘Muller Thurgau’ sensitive to downy mildew were used as controls in the leaf disk assay.
DNA extraction
The extraction of plant DNA from the investigated varieties was performed with Qiagen_DNeasy 96 Plant Kit (Qiagen GmbH, Hilden, Germany). Sporangia for DNA extraction were collected from the infected leaves in filter tips using a vacuum pump. The collected material was frozen at −20 °C. Extraction of DNA from sporangia was carried out with innuPREP plant DNA kit (Analytik Jena AG, Jena, Germany).
Microsatellite analysis
Nine microsatellite markers adopted by the GrapeGen06 (http://www.montpellier.inra.fr/grapegen06) project for genetic identification of grapevines were used for microsatellite analysis of the grape cultivars: VVMD5, VVMD7, VVMD25, VVMD27, VVMD28, VVMD32, VVS2, ssrVrZag62 and ssrVrZag79 [Citation14–17]. Polymerase chain reaction (PCR) conditions were set according to Dzhambazova et al. [Citation18]. Forward primers of markers were labelled with 6-FAM, ATTO565, ATTO550 and Yakima Yellow florescent dyes.
For microsatellite profiling of different P. viticola isolates, the following seven microsatellite loci were used: Pv65, Pv101, Pv137, Pv140, Pv143, Pv144 and Pv147 [Citation8]. PCR reaction and cycles were performed as described in Rouxel et al. [Citation8]. The forward primers of all markers were fluorescently labelled with 6-FAM with the exception of Pv140, which was labelled with ATTO656.
The SSR markers used for detection of resistance-related alleles at the Rpv3 locus were UDV305 and UDV737 [Citation2]; at the Rpv12 locus, UDV340 and UDV014 [Citation19]; and at locus Rpv10, Gf09-46 and GF09-55 [Citation6]. PCR conditions were as described in the corresponding publications. All forward primers were fluorescently labelled with 6-FAM with the exception of UDV340, which was labelled with ATTO656.
Fragment analysis of the amplified products was carried out on an ABI 3130 Genetic Analyzer (Applied Biosystems, Foster city, CA, USA). For calculation of allele sizes, Orange 500 DNA Size Standard (500 bp) (MCLAB, San Francisco, CA, USA) was used. Alleles were scored using the Genemapper v.4.0 software (Applied Biosystems).
Data analysis
GenAlex version 6 [Citation20] was used for calculation of allele frequencies, expected and observed heterozygosity. One-way analysis of variance in MS Excel was used for determination of: (1) the genotypic effect on the level of resistance for each isolate and (2) the effect of a particular isolate on the level of resistance of each variety.
Results and discussion
Genotyping of grapevine varieties
Eighteen Bulgarian newly bred varieties, obtained by interspecific crossing, were genotyped with nine microsatellite markers adopted for genetic identification of grapevines by the GrapeGen06 project (http://www.montpellier.inra.fr/grapegen06). As a result, 18 unique genotypes were determined. The microsatellite profiles of the studied cultivars are shown in Table S1 (Online Supplementary Appendix).
Considering the pedigree of the investigated varieties, they were analysed for the presence of three Rpv loci, Rpv3, Rpv10 and Rpv12, underlying the resistance to P. viticola. Varieties ‘Bianca’, ‘Kunbarat’ and V. amurensis Ruprecht were also included in the analysis as controls for the presence of resistance-related alleles of the Rpv3, Rpv12 and Rpv10 loci, respectively.
The Rpv3 locus conferring resistance to P. viticola [Citation21] is flanked by the SSR markers UDV305 and UDV737.[Citation2] Resistance-related alleles at these markers, 300 bp at UDV305 and 281 bp at UDV737, were found in 17 out of the 18 analysed varieties as well as in the referent variety ‘Bianca’ (). These resistance-related alleles have been inherited from Villard Blanc, which is known to be a parent or grandparent in the pedigree of these 17 varieties as well as in the referent variety ‘Bianca’ [Citation1]. Villard Blanc is a complex hybrid, whose resistance originates from its resistant ancestors, five North American Vitis species: V. aestivalis, V. berlandieri, V. cinerea, V. lincecumii and V. rupestrisin. The haplotype Rpv3299-279 of SV12375 at markers UDV305 and UDV737 has been found to be the most represented haplotype in the breeding lines and resistant cultivars, including ‘Bianca’, from the North American ancestry [Citation2]. Thus, we can conclude that, in agreement with their parentage, 17 of the analysed varieties shared the Rpv3 haplotype of Villard Blanc.
The investigated sets of varieties were analysed with markers GF09-46 and GF09-55 linked to the Rpv10 locus [Citation6]. The resistance-related alleles for both markers, 411 bp for GF09-46 and 250 bp for GF09-55, were found only in cultivar ‘Slava’ and the accession of V. amurensis Ruprecht (). This is in agreement with the pedigree of cv. ‘Slava’, which is a descendant of V amurensis. ‘Slava’ was created by a cross between ‘Dunavska Gamza’ and ‘Tsvetochnyi’. The parent of ‘Tsvetochnyi’ is ‘Severnyi’, which was obtained by a cross between ‘Seyanets Malengra’ and V. amurensis [Citation1]. It harbours Rpv10 [Citation2,6] inherited from V. amurensis [Citation2]. Thus, Rpv10 has been transmitted from V. amurensis to cultivar ‘Slava’ through its grandparent Severnyi and parent ‘Tsvetochnyi’.
The analysed varieties as well as the control variety ‘Kunbarat’ were further genotyped with marker UDV340, which co-segregates with Rpv12, and marker UDV014, which flanks the Rpv12 locus [Citation19]. ‘Kunbarat’ (V. amurensis × V. vinifera) × ‘Italia’) [Citation1] was used in the analysis as a referent cultivar, because it harbours Rpv12 [Citation19]. The presence of Rpv12 resistance-related allele of 181 bp at marker UDV340 was found in ‘Droujba’, ‘Garant’, ‘Kajlushki Rubin’, ‘Plevenska rosa’ and ‘Kunbarat’ (). These varieties, with the exception of ‘Kajlashki rubin’ and ‘Kunbarat’, are descendants of ‘Zarya Severa’, which is a cross between ‘Seyanets Malengra’ and V. amurensis [Citation1] and harbours Rpv12 originating from V. amurensis [Citation19]. According to the recorded pedigree, ‘Zarya Severa’ is a grandparent of ‘Droujba’, which is, on the other hand, a parent of ‘Garant’ and ‘Plevenska rosa’ (). It seems that during the crossing process, Rpv12 from ‘Zarya Severa’ has been transmitted to ‘Garant’ and ‘Plevenska rosa’ through ‘Droujba’.
The Rpv12 haplotype at marker UDV014 has an allele size of 160 bp [Citation19]. Interestingly, this haplotype is shared by ‘Droujba’, ‘Garant’, ‘Plevenska rosa’ and ‘Kunbarat’, but not by Kajlushki Rubin (). Probably, mutation or crossover is the possible reason for the lack of resistance-related allele at this marker in ‘Kajlushki Rubin’.
Genotypig of P. viticola isolates
The samples of seven P. viticola isolates originating from seven different regions of Bulgaria were genotyped with seven SSR markers developed by Rouxel et al. [Citation8]. The results from the calculated genetic diversity parameters are shown in . The number of alleles per locus varied between 1 and 6 with a mean value of 3, while the mean value of effective alleles was 2.26 per locus. The results for the observed heterozigocity varied between 0 and 1 with a mean value of 0.694 ± 0.165, which is higher from the mean value of expected heterozigocity (0.462 ± 0.107). The genetic polymorphism revealed by the used microsatellite markers allowed the discrimination of all seven P. viticola isolates.
Table 2. Genetic diversity parameters found in the set of seven P. viticola isolates.
Assessment of the level of downy mildew resistance
The assessment of the level of resistance to downy mildew of the investigated set of varieties was performed with seven different P. viticola isolates. Cultivars ‘Kunbarat’ and ‘Mueller Thurgau Weiss’ were used in the leaf disk assay as controls for high and low level of resistance to downy mildew, respectively. The analysis of variance performed for each P. viticola isolate showed significant differences (P < 0.05) between the investigated varieties.
The effect of the particular isolate on the level of resistance was evaluated for each variety. The results showed a clear correlation between the presence of different Rpvs and the resistance scores recorded for the corresponding cultivar. The OIV452 rates of five varieties that contain Rpv10/Rpv3 (‘Slava’), Rpv12/Rpv3 (‘Droujba’, ‘Garant’, Plevensaka Rosa) or only Rpv12 (‘Kajlashki rubin’) varied between 5 and 9 with mean values of 6.5–8.25 among the different isolates, while the rate of the remaining varieties, harbouring only Rpv3 as well as ‘Bianca’ varied in a broad range of OIV452 scores from 1 to 5 with a mean value of 1.75–4.8 (). These results demonstrated that all the seven isolates used for inoculation in the leaf disk assay were able to infect the varieties harbouring only Rpv3, thus overcoming the resistance determined by Rpv3. This finding is not surprising in view of the results that particular P. viticola isolates can evade the resistance conferred by Rpv3 in ‘Bianca’ Citation[3,19]. In contrast, the presence of Rpv10 and Rpv12 ensured the maintenance of high levels of resistance over all P. viticola isolates used for infection. These results are in accordance with those reported by Schwander et al. Citation[6] and Venuti et al. Citation[19] for high levels of resistance conferred by Rpv10 and Rpv12. The OIV452 values of ‘Droujba’, ‘Garant’ and ‘Plevenska rosa’, which harbour Rpv12 and Rpv3, were comparable over the seven isolates of P. viticola with those of ‘Kajlashki rubin’, which has only Rpv12. Thus, the additive effect of two Rpvs in one genotype (Rpv12/Rpv3 or Rpv10/Rpv3) found in the study of Venuti et al. Citation[19] and Schwander et al. Citation[6] was not observed in our investigation. Probably other resistance factors in the genome of ‘Kajlashki rubin’, which were inherited from V. amurensis, but were not analysed in this study, are responsible for the high level of resistance against downy mildew observed for this cultivar. Further investigations are necessary in order to determine the resistance factors which are presented in this variety.
Figure 1. Distribution of OIV 452 values of investigated varieties over seven P. viticola isolates. Note: The numbers at outliers indicate the number of the corresponding P. viticola isolate: Galabnik (1), Dobromirka (2), Pleven (3), Pomorie (4), Ruse (5), Sofia (6) and Vidin (7).
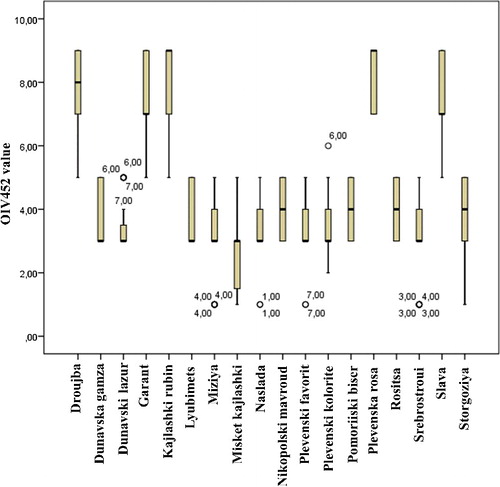
Overall, the performed phenotypic and molecular analysis proved that the high level of resistance to downy mildew against seven P. viticola isolates obtained in ‘Droujba’, ‘Garant’ and ‘Plevenska rosa’, ‘Kajlashki rubin’ and ‘Slava’ corresponds to the presence of Rpv10 and Rpv12 in the genome of these cultivars, while a quite lower level of resistance was observed in all cultivars carrying only Rpv3.
Conclusions
Here, we presented the characterization of 18 Bulgarian newly bred varieties obtained by interspecific crossing, in regard with their genetic identity, the level and the stability of their resistance to downy mildew against seven different isolates of P. viticola as well as the presence of the genetic loci conferring resistance to downy mildew in these varieties. The performed molecular analysis revealed, in accordance with the parentage of the investigated varieties, the presence of Rpv loci of different origin, North American Vitis species and V. amurensis. A clear correlation between the presence of particular Rpv and the level and the stability of the resistance was determined. Five cultivars which harbour resistance factors from V. amuersis, Rpv10 or Rpv12, alone or in a combination with Rpv3, were able to maintain high level of resistance to all seven P. viticola isolates. The lower level of resistance of the cultivars that contain only Rpv3 inherited from cultivar Villard Blanc was found to vary significantly among the different isolates. Based on the results obtained in this study, we could recommend five grapevine varieties with broad and durable resistance and good quality of grape and wine, ‘Droujba’, ‘Garant’ and ‘Plevenska rosa’, ‘Kajlashki rubin’ and ‘Slava’, as a valuable material for further MAS selection and pyramiding of different resistance loci.
Supplementary_Data.pdf
Download PDF (229.7 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Vitis International Variety Catalogue [ Internet]. Quedlinburg: Julius Kühn-Institut – Federal Research Centre for Cultivated Plants (JKI); Siebeldingen: Institute for Grapevine Breeding – Geilweilerhof (ZR); c2016 [ updated 2016 Aug; cited 2016 Oct 28]. Available from: http://www.vivc.de
- Di Gaspero G, Copetti D, Coleman C, et al. Selective sweep at the Rpv3 locus during grapevine breeding for downy mildew resistance. Theor Appl Genet. 2012;124:277–286.
- Peressotti E, Wiedemann-Merdinoglu S, Delmotte F, et al. Breakdown of resistance to grapevine downy mildew upon limited deployment of a resistant variety. BMC Plant Biol [ Internet]. 2010 [ cited 2016 Oct 28];10:147. Available from: http://bmcplantbiol.biomedcentral.com/articles/10.1186/1471-2229-10-147.
- Eibach R, Zyprian E, Welter L, et al. The use of molecular markers for pyramiding resistance genes in grapevine breeding. Vitis. 2007;46:120–124.
- Töpfer R, Hausmann L, Eibach R. Molecular breeding. In: Adam-Blondon AF, Martinez-Zapater JM, Kole C, editors. Genetics, genomics and breeding of grapes. Enfield: Science Publishers; 2011. p. 160–185.
- Schwander F, Eibach R, Fechter I, et al. Rpv10: a new locus from the Asian Vitis gene pool for pyramiding downy mildew resistance loci in grapevine. Theor Appl Genet. 2012;124:163–176.
- Delmotte, F., W. Chen, S. Richard-Cervera, et al. Microsatellite DNA markers for Plasmopara viticola, the causal agent of downy mildew of grapes. Mol Ecol Notes. 2006;6:379–381.
- Rouxel M, Papura D, Nogueira M, et al. Microsatellite markers for characterization of native and introduced populations of Plasmopara viticola, the causal agent of grapevine downy mildew. Appl Environ Microbiol. 2012;78(17):6337–6340
- Ivanov M, Simeonov I, Nakov Z. ‘Garant’ – new table grapes interspecies variety. Ann Univ Craiova– Horti Food Prod Processing Technology Series. 2015;20:193–198. Bulgarian.
- Ivanov M, Nakov Z, Simeonov I, et al. Kaylashki Rubin – a new red wine grapevine variety. Agr Sci. 2011;44:60–65. Bulgarian.
- Ivanov M, Nakov Z, Simeonov I. ‘Plevenska rosa’ – a new white wine grapevine variety. Agr Sci. 2011;44:80–85. Bulgarian.
- Valchev V, Ivanov Y, Petkov G, et al. Slava – new wine grape variety. Viticult Enol. 1992;6:9. Bulgarian.
- OIV descriptor list for grape varieties and Vitis species. 2nd ed. Paris: Office International de la Vigne et du Vin; 2009. Available from: http://www.oiv.org
- Thomas M, Scott N. Microsatellite repeats in grapevine reveal DNA polzmorphisms when analysed as sequence-tagged sites (STSs). Theor Appl Genet. 1993;86;985–990.
- Bowers J, Dangl G, Vignani R, et al. Isolation and characterization of 345 new polymorphic simple sequence repeat loci in grape (Vitis vinifera L.). Genome. 1996;39:628–633.
- Bowers J, Dangl G, Meredith C. Development and characterization of additional microsatellite DNA markers for grape. Am J Enol Viticult. 1999;50:243–246.
- Sefc K, Regner F, Turetschek E, et al. Identification of microsatellite sequences in Vitis riparia and their applicability for genotyping of different Vitis species. Genome. 1999;42:367–373.
- Dzhambazova T, Tsvetkov I, Simeonov I, et al. Genetic diversity and relationship of indigenous and newly bred Bulgarian grape cultivars assessed by nuclear and chloroplast markers. J Int Des Sci Vigne Vin. 2012;46:113–121.
- Venuti S, Copetti D, Foria S, et al. Historical introgression of the downy mildew resistance gene Rpv12 from the Asian species Vitis amurensis into grapevine varieties. PLoS One [Internet]. 2013 [ cited 2016 Oct 28];8(4):e61228. Available from: http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0061228
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295.
- Bellin D, Peressotti E, Merdinoglu D, et al. Resistance to Plasmopara viticola in grapevine ‘‘Bianca’’ is controlled by a major dominant gene causing localised necrosis at the infection site. Theor Appl Genet. 2009;120:163–176.
