ABSTRACT
In this study, inter-simple sequence repeats (ISSR) and start codon targeted (SCoT) markers were used for genetic diversity and relationship analysis of nine Salvia species. Twenty-one and twenty selected ISSR and SCoT primers amplified 350 and 329 loci, respectively, of which all were polymorphic. The obtained average polymorphism information content (ISSR, 0.38; SCoT, 0.40), average band informativeness (ISSR, 16.67; SCoT, 16.45) and resolving power (ISSR, 9.75; SCoT, 12.52) revealed high genetic diversity prevailing among Salvia accessions. Considering the ISSR and SCoT data, the species with a basic chromosome number of x = 8 showed higher values of the percentage polymorphism loci (PPL), the number of observed alleles (Na) and Shannon index (I) than the other species. The partition of clusters in the neighbour-joining dendrogram based on ISSR, SCoT and combined data was similar and grouped all individuals into four clusters. However, the dendrogram generated based on SCoT separated the individuals into sub-clusters in accordance with their species and section. The Mantel test revealed a similar polymorphism distribution pattern between ISSR and SCoT techniques, the correlation coefficient (r) was 0.83, and the results showed that both techniques were effective to assess the genetic diversity. Our results indicated that SCoT markers can be used as a reliable and informative technique for evaluation of genetic diversity and relationships among Salvia species.
Introduction
Salvia (Garden sage) is one of the most important genera in the Labiatae family and includes nearly 1000 species all over the world [Citation1]. Many Salvia species are used as herbal tea and in cosmetics, perfumes and the pharmaceuticals [Citation2]. Furthermore, plants from this genus are distinguished for their biological activities such as antioxidant, antitumour, anti-inflammatory, antimicrobial, antidiabetic and anxiolytic [Citation3–6]. Mediterranean and western Asia areas have been considered as the original centers of the distribution of this genus, and the Iranian flora includes 58 Salvia species, of which 17 are endemic to Iran [Citation7].
With the progress in plant molecular biology, numerous molecular marker techniques have been developed and used widely in evaluating genetic diversity, population structure and phylogenetic relationships. In recent years, advances in genomic tools provide a wide range of new marker techniques such as, functional and gene-targeted markers as well as develop many novel DNA-based marker systems [Citation8]. Start codon targeted (SCoT) polymorphism is one of the novel, simple and reliable gene-targeted marker systems. This molecular marker offers a simple DNA-based marker alternative and reproducible technique which is based on the short conserved region in the plant genes surrounding the ATG [Citation9] translation start codon [Citation10]. This technique involves a polymerase chain reaction (PCR) based DNA marker with many advantages such as low-cost, high polymorphism and extensive genetic information [Citation11]. The SCoT system has been successfully used to assess genetic diversity, carry out structure analysis, identify cultivars, map quantitative trait loci (QTL), as well as perform DNA fingerprinting and diagnosis in different species, including peanut [Citation12], check pea [Citation13], potato [Citation14], orchids [Citation15], coconut [Citation16] and wheat [Citation17,Citation18].
So far, there have been few attempts to study the genetic variation in Salvia species using different molecular markers [Citation19–21]. However, no studies have been conducted to assess the genotypic differences and relationships in Salvia species using gene-targeted markers, and this is the novelty of the current work. Therefore, the objectives of this study are:
to consider and compare polymorphic SCoT and ISSR techniques with highly informative values and to characterize the genetic variability in selected accessions;
to evaluate genetic relationships among wild accessions and specifically to measure the extent of variation between and within ploidy and chromosomal groups.
Materials and methods
Plant material and DNA extraction
A total of 20 accessions of Salvia belonging to nine species (S. aethiopis, S. macrosiphon, S. nemorosa, S. officinalis, S. reuterana, S. sclarea, S. virgata, S. verticillata and S. syriaca) were collected from different geographical regions of Iran. Additional information about the sampling locations, ploidy level, genus and chromosomal groups are shown in . Genomic DNA was isolated from young leaves following the CTAB (cetyl-trimethyl-ammonium bromide) protocol [Citation22]. The quality and quantity of the extracted DNA were checked by 0.8% agarose gel electrophoresis.
Table 1. Passport of Salvia accessions used in this study.
ISSR-PCR amplification
For the ISSR analysis, 31 primers from UBC (University of British Columbia) series were tested for DNA amplification. Twenty-one primers were chosen for ISSR analysis of genetic variability, based on band reproducibly (). The total reaction volume was 20 µL containing 10 µL master mix 2XPCR (ready-to-use PCR master mix 2X), 2 µL of isolated DNA from each accession, 2 µL of each primer and 6 µL ddH2O (double distilled water). The PCR amplification (BioRad, T-100) was run at 94 °C for 6 min, followed by 35 cycles of denaturation at 94 °C for 30 s, primer annealing at 46.9–60.5 °C (varied for each primer) for 45 s and primer elongation at 72 °C for 2 min. The final extension was 7 min at 72 °C. The amplification reaction products were detected by 1.5% denaturing agarose gels stained with safe view ΙΙ.
Table 2. Details about the banding pattern revealed by ISSR and SCoT primers.
SCoT-PCR amplification
A total of 20 SCoT primers used in this study were designed according to Collard and Mackill [Citation10] and shown in . The PCRs were done in 20-µL reaction mixtures containing10 µL master mix 2XPCR (ready-to-use PCR master mix 2X), 6 µL ddH2O, 2 µL of isolated DNA from each sample and 2 µL of each primer. Amplification was run at 94 °C for 5 min, followed by 45 cycles of denaturation at 94 °C for 45 s, primer annealing at 45 °C for 45 s and primer elongation at 72 °C for 90 s. The final extension was 10 min at 72 °C. The amplification reaction products were detected by 1.5% denaturing agarose gels stained with safe view ΙΙ.
Data analysis
Firstly, amplification products were scored as absent (0) and present (1), compiling the data as a binary matrix. For comparing the banding patterns of primers, various genetic parameters such as polymorphism information content (PIC) [Citation23], resolving power (Rp) [Citation24] and marker index (MI) [Citation25] were calculated. To detect the distribution of genetic variation within and among groups, analysis of molecular variance (AMOVA) was done using GenAlEx software [Citation26]. The genetic variation indices, viz. the percentage of polymorphic loci (PPL), the observed (Na) and effective numbers of alleles (Ne), Nei's gene diversity (H), and Shannon's information index (I) were estimated using POP-GENE software [Citation27]. The genetic similarities matrix from the calculative data was used to construct a dendrogram based on the neighbour-joining (NJ) method using DARwin software [Citation28]. In addition, the correlations among ISSR and SCoT matrices were measured using Mantel's test [Citation29] statistics using XLSTAT package.
Results and discussion
Polymorphism detected by ISSR and SCoT primers
Twenty-one ISSR primers amplified a total of 350 scorable bands (), of which all were polymorphic, accounting for 100%. The number of polymorphic bands ranged from 11 (ISSR-26) to 21 (ISSR-14), with an average of 16.67 fragments per primer. The resolving power (Rp) of the primers varied from 4.90 (by ISSR-2) to 13.30 (by ISSR-19), with an average of 9.75. The highest value of the marker index (MI) was obtained with primer ISSR-4 (8.28), while the ISSR-26 primer showed the lowest value (3.16). The polymorphism information content (PIC) of the primers ranged from 0.28 (by ISSR-7) to 0.45 (by ISSR-3) with an average of 0.38 per primer.
With the SCoT system, a total of 329 scorable bands were amplified using the 20 primers, of which all were polymorphic, with an average of 16.45 polymorphic bands per primer. The number of polymorphic bands ranged from 11 (SCoT-17 and SCoT-26) to 24 (SCoT-20) with an average of 16.45. The average percentage of PIC was 40 and also the highest value of this index (0.47) was obtained with primers SCoT-1 and SCoT-15. The highest Rp (16.1) and MI (9.75) values were obtained with primers SCoT-15 and SCoT-20, respectively. Further details of the primers are given in .
DNA analysis using ISSR and SCoT markers has proved to be an efficient and inexpensive way to provide molecular data to evaluate genetic variation, and it has been used successfully to determine genetic relationships for many plants [Citation12,Citation14,Citation17,Citation18,Citation30–33]. In the present study, the ISSR technique revealed higher mean PB (16.67) than SCoT (16.45). However, the mean values of Rp, MI and PIC were higher for SCoT than for ISSR (). Of these, the polymorphism information content (PIC) and resolving power (Rp) provide a degree of capability of marker systems, which help to determine the effectiveness and potential of the primers used in the fingerprinting process [Citation25]. In other words, PIC is the probability of detection of polymorphism by a primer/primer combination between two randomly drawn individuals and depends on the number of detectable alleles and the distribution of their frequency. Moreover, another important feature of a good marker technique is the capability to discriminate among different individuals. Rp specifies the discriminatory potential of the primers. In fact, this parameter estimates the ability of a primer or technique to produce optimally informative bands, which were calculated per individual for each primer to determine its efficiency [Citation34]. Our results showed that the average values of PIC and Rp of SCoT primers are more than those of ISSR primers, which illustrates the good applicability of the SCoT technique in the assessment of genotyping in Salvia species. Our findings are in agreement with Gorji et al. [Citation14], Rajesh et al. [Citation16], Etminan et al. [Citation17] and Pour-Aboughadareh et al. [Citation18,Citation33], who demonstrated the usefulness of SCoT markers for estimation of genetic diversity and fingerprinting of genotypes more than other DNA marker systems.
Genetic diversity analysis
Using the ISSR technique, 91 and 86% of the genetic variation were partitioned within ploidy levels and the basic chromosomes groups. Based on the results from the SCoT technique and combined data (ISSR + SCoT), the AMOVA revealed that 87 and 81%, and 89 and 84% of the genetic variation were partitioned within ploidy groups and the basic chromosome groups, respectively (). In general, these results revealed that there exists a higher distribution of genetic diversity within groups compared to among groups.
Table 3. Analysis of molecular variance (AMOVA) based on ISSR and SCoT markers for Salvia species.
A summary of genetic variation parameters is given in . As shown in this table, using both ISSR and SCoT techniques, the highest values for the percentage of polymorphic loci, the number of observed alleles (Na) and Shannon index value (I) were observed for accessions with eight basic chromosomes (PPL = 92.57%, Na = 1.85 and I = 0.37). Additionally, based on ISSR primers, diploid species showed the highest values of the maximum values for PPL (71.71%), Na (1.46), I (0.37), the number of effective alleles (Ne = 1.41) and Nei's gene diversity (H = 0.24). According to SCoT primers, the highest values of Ne, H and I were those of the tetraploid accessions. However, the diploid species showed the highest values for the PPL and Na indices.
Table 4. Genetic variation features estimated using ISSR and SCoT markers in the studied Salvia species.
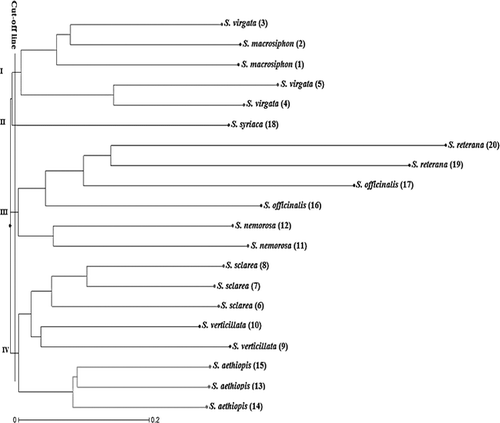
There were some differences in the genetic similarity (GS) among individuals determined by both ISSR and SCoT techniques (data not shown). For the ISSR markers, the GS-based pairwise comparisons indicated that the genetic similarity varied from 0.01 (between S. macrosiphhon and S. verticillata) to 0.61 (between two S. sclarea individuals). On the other hand, the GS values obtained by SCoT markers indicated that two S. officinalis individuals collected from Hamadan and Esfahan are relatively closest with a coefficient of 0.63, whereas S. virgata and S. aethiopis are the most distant individuals with a similarity coefficient of 0. However, the Mantel matrix correspondence test indicated a good association between them. The correlation between GS values based on these markers (r = 0.83) was significantly high, revealing a great correspondence of polymorphisms brought out by these marker techniques. In this regard, Rahmani et al. [Citation35] also showed a significant correlation between SCoT and ISSR markers. The neighbour-joining (NJ) dendrogram generated by ISSR, SCoT and combined data (ISSR + SCoT) separated the 20 individuals of Salvia into four main clusters (). In all dendrograms, the individuals were grouped according to their species and were separated in the same sub-clusters. Although part of the individuals were consistently clustered together in all three cluster analyses, the dendrogram generated based on SCoT markers showed a clear clustering pattern of genetically closer individuals. For instance, as shown in , the first main cluster divided into two sub-clusters; sub-cluster I consisted of S. nemorosa and S. virgata, and sub-cluster II included S. sclarea species. The second cluster also divided into two sub-clusters so that S. athiophisand S. verticillata separated from each other. The third cluster included S. officinalis, S. reuterana and S. macrosiphon individuals, and S. syriaca a lonely generated forth cluster. On the other hand, in the dendrogram obtained by ISSR, two individuals related to S. officinalis separated in two different sub-clusters.
Figure 1. Dendrogram of Salvia accessions based on ISSR data using the neighbour-joining (NJ) method.
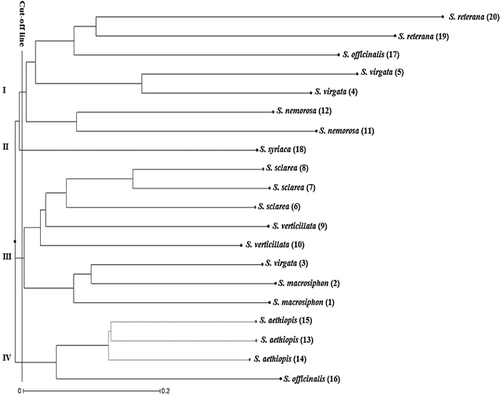
Figure 2. Dendrogram of Salvia accessions based on SCoT data using the neighbour-joining (NJ) method.
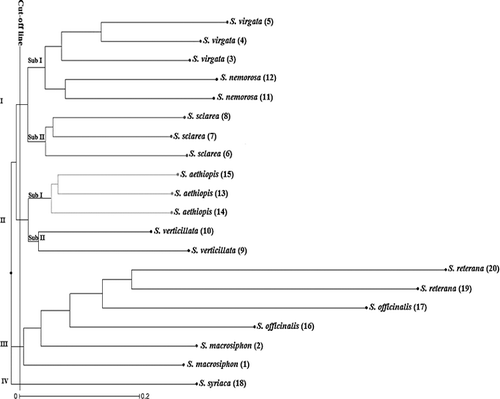
Figure 3. Dendrogram of Salvia accessions based on combined data (ISSR + SCoT) using the neighbour-joining (NJ) method.
The present study, based on two genetic marker systems, revealed high genetic variability within Salvia populations. Hence, identification, collection, as well as conservation of Salvia species from different geographical regions of Iran, would be of great importance in the formulation of maintenance strategies for the species belonging to this genus. In line with these results, some researchers showed high levels of genetic diversity in several species of genus Salvia [Citation19–21]. The potential of SCoT markers for analyzing the genetic diversity and genetic relationships among Salvia species is another key result with practical implications. Thus, we supported the available evidence that this molecular technique could either be utilized individually or in combination with other molecular techniques to evaluate the genetic variation and to obtain reliable information about genetic relationships [Citation8], which would aid strategies for effective collection of Salvia germplasm and their conservation.
Conclusions
Our findings revealed the existence of more genetic variability in Iranian wild Salvia populations. More importantly, the results revealed that the SCoT technique could be used for assessment of genetic relationships, which ultimately would be helpful in differentiation of various other representatives of the Salvia genus.
Acknowledgement
This article is published based on a research supported by vice chancellor of research and technology of Kermanshah Branch, Islamic Azad University. The authors acknowledge with thanks the technical and lab facilities support from the Plant Biotechnology Research Center of Kermanshah Branch, Islamic Azad University, Kermanshah, Iran.
Disclosure statement
The authors declare there is no conflict of interest for their study.
References
- Longaray-Delamare AP, Moschen-Pistorello IT, Artico L, et al. Antibacterial activity of the essential oil of Salvia officinalis L. and Salvia triloba L. cultivated in South Brazil. Food Chem. 2007;100:603–608.
- Coisin M, Laudia-Padurariu C, Raluca-Andro A, et al. Biochemical and physiological researches in Salvia nemorosa L. An Stiint Univ Al I Cuza Iasi Sect II a Biol veget. 2012;58:51–58.
- Kelen M, Tepe B. Chemical composition, antioxidant and antimicrobial properties of the essential oils of three Salvia species from Turkish flora. Bioresour Technol. 2008;99:4096–4104.
- Cardilea V, Russob A, Formisanoc C, et al. Essential oils of Salvia bracteata and Salvia rubifolia from Lebanon: chemical composition, antimicrobial activity and inhibitory effect on human melanoma cells. J Ethno Pharmacol. 2009;126:265–272.
- Akin M, Demirci B, Bagci Y, et al. Antibacterial activity and composition of the essential oils of two endemic Salvia sp. from Turkey. Afr J Biotechnol. 2010;9:2322–2327.
- Orhan EI, Senol SF, Ercetin T, et al. Assessment of anticholinesterase and antioxidant properties of selected sage (Salvia) species with their total phenol and flavonoid contents. Ind Crops Prod. 2013;41:21–30.
- Rechinger KH. Flora Iranica. Wien, (Austria): Akademische Druke-U. Verlagsanstalt; 1963.
- Poczai P, Varga I, Laos M, et al. Advances in plant gene-targeted and functional markers: a review. Plant Methods. 2013;9:6–37.
- Sawant SV, Singh PK, Gupta SK, et al. Conserved nucleotide sequences in highly expressed genes in plants. J Genet. 1999;78:123–131.
- Collard BCY, Mackill DJ. Start Codon Targeted (SCoT) Polymorphism: a simple, novel DNA marker technique for generating gene-targeted markers in plants. Plant Mol Biol Report. 2009;27:86–93.
- Guo DL, Zhang JY, Liu CH. Genetic diversity in some grape varieties revealed by SCoT analyses. Mol Biol Rep. 2012;39:5307–5313.
- Xiong F, Zhong R, Han, Z, et al. Start codon targeted polymorphism for evaluation of functional genetic variation and relationships in cultivated peanut (Arachis hypogaea L.) varieties. Mol Biol Rep. 2011;38:3487–3494.
- Amirmoradi B, Talebi R, Karami E. Comparison of genetic variation and differentiation among annual Cicer species using start codon targeted (SCoT) polymorphism, DAMD-PCR, and ISSR markers. Plant Syst Evol. 2012;298:1679–1688.
- Gorji AM, Poczai P, Polgar Z, et al. Efficiency of arbitrarily amplified dominant markers (SCoT, ISSR and RAPD) for diagnostic fingerprinting in tetraploid potato. Am Potato J. 2011;88:226–237.
- Bhattacharyya P, Kumaria S, Kumar S, et al. Start codon targeted (SCoT) marker reveals genetic diversity of Dendrobium nobile Lindl., an endangered medicinal orchid species. Gene. 2013;529:21–26.
- Rajesh MK, Sabana AA, Rachana KE, et al. Genetic relationship and diversity among coconut (Cocos nucifera L.) accessions revealed through SCoT Analysis. 3 bitech. 2015;5:999–1006.
- Etminan A, Pour-Aboughadareh A, Mohammadi R, et al. Applicability of start codon targeted (SCoT) and inter-simple sequence repeat (ISSR) markers for genetic diversity analysis in durum wheat genotypes. Biotechnol Biotechnol Equip. 2016;30:1075–1081.
- Pour-Aboughadareh A, Ahmadi J, Mehrabi AA, et al. Insight into the genetic variability analysis and relationships among some Aegilops and Triticum species, as genome progenitors of bread wheat, using SCoT markers. Plant Biosyst. Forthcoming 2018. DOI:10.1080/11263504.2017.1320311.
- Erbano M, Schuhli GS, Santos EP. Genetic variability and population structure of Salvia lachnostachy: implications for breeding and conservation programs. Int J Mol Sci. 2015;16:7839–7850.
- Safaei M, Sheidai M, Alijanpoor, et al. Species delamination and genetic diversity analysis in Salvia with the use of ISSR molecular markers. Acta Bot Croat. 2016;75:45–52.
- Yousefiazar-Khanian M, Asghari A, Ahmadi J, et al. Genetic diversity of Salvia species assessed by ISSR and RAPD markers. Not Bot Horti Agrobo. 2016;44:431–436.
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15.
- Bootstein D, White RL, Skolnick M, et al. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980;32:314–331.
- Prevost A, Wilkinson MJ. A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genet. 1999;98:107–112.
- Powell W, Morgante M, Andre C, et al. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed. 1996;2:225–238.
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Resour. 2006;6:288–295.
- Yeh FC, Yang RC, Boyle T. Microsoft Window-based freeware for population genetic analysis (POPGENE). Version 1.31. 1999. Available from: http://ftp.microsoft.com/softlib/MSLFILES/HPGL.EXE.
- Perrier X, Flori A, Bonnot F. Data Analysis Methods. In: Hamon P, Seguin M, Perrier X, Glaszmann JC, editors. Genetic diversity of cultivated tropical plants. Boca Raton, FL (USA): CRC Press; 2003. p. 360.
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220.
- Hajibarat Z, Saidi A, Hajibarat Z, et al. Characterization of genetic diversity in chickpea using SSR markers, start codon targeted polymorphism (SCOT) and conserved DNA-derived polymorphism (CDDP). Physiol Mol Biol Plants. 2015;21:365–373.
- Moradkhani H, Mehrabi AA, Etminan A, et al. Molecular diversity and phylogeny of Triticum-Aegilops species possessing D genome revealed by SSR and ISSR markers. Plant Breed Seed Sci. 2015;71:82–95.
- Pour-Aboughadareh A, Mohmoudi AM, Ahmadi J, et al. Agro-morphological and molecular variability in Triticum boeoticum accessions from Zagros Mountains, Iran Genet Resour Crop Evol. 2017;64:545–556.
- Pour-Aboughadareh A, Ahmadi J, Mehrabi AA, et al. Assessment of genetic diversity among Iranian Triticum germplasm using agro-morphological traits and start codon targeted (SCoT) markers. Cereal Res Commun. 2017;45:574–586.
- Yousefi V, Najaphy A, Zebarjadi A, et al. Molecular characterization of Thymus species using ISSR markers. J Anim Plant Sci. 2015;25:1087–1094.
- Rahmani MS, Alikhani L, Shabanian N, et al. Genetic differentiation in Quercus infectoria from northwest of Iran revealed by different nuclear markers. Tree Genet Genomes. 2015;11:800–809.
