ABSTRACT
IL-33 binding to IL1RL1 is involved in asthma. The data show that involvement of the IL33/IL1RL1 genes in asthma is biologically plausible. The objective of this study was to investigate the association of IL33 and IL1R1 gene polymorphisms and serum IL-33 levels with asthma within the population of Xinjiang (China). A case group of 279 patients and 277 healthy controls were genotyped using the MassARRAY SNP genotyping system. In 113 asthma patients and 112 controls, the IL-33 levels were measured by enzyme linked immunosorbent assay (ELISA). The statistical analysis showed that IL-33 levels were associated with asthma and the rs16924159 (AA), rs1420102 (TT) genotype was associated with a significantly increased risk of asthma compared with the GG (rs16924159) or CC (rs1420102) genotypes. However, neither the rs13431828C/T genotypes, nor the rs928413 A/G genotypes were significantly associated with risk of asthma. In addition, the patients carrying the rs16924159 AA genotype presented with higher IL-33 levels compared to the GG group. This result suggested that the increase of IL-33 in patients with asthma may affect the expression level of serum IL-33. Furthermore, the rs16924159 G-A variant was associated with IL-33 levels in patients with asthma.
Introduction
Asthma is characterised by recurrent episodes of airway obstruction, hyperproduction of immunoglobulin E (IgE), mucus hypersecretion and hyperresponsiveness, which affect more than 300 million people globally, and is usually associated with chronic airway inflammation [Citation1,Citation2]. Accumulating evidence has shown that asthma is a multifactorial disease, which results from complex interactions between a variety of genetic and environmental factors [Citation3,Citation4], but its detailed mechanism has not been fully elucidated yet.
In recent years, Interleukin 33 (IL-33) has been reported as a new member of the IL-1 family of cytokines, with the ability to induce Th2 cytokine production and amplify both Th1 and Th2-type responses [Citation5–7]. The functions of IL-33 are mediated through its receptor, IL-1 receptor like 1 (IL1RL1), and the IL-1 receptor accessory protein (IL1RAcP) [Citation8]. This receptor complex induces, through activation of signalling proteins, release of allergic and eosinophilic mediators, such as IL-5 and IL-13, resulting in eosinophilic inflammation and attenuation of the IL-33 signal [Citation9,Citation10]. These data show that involvement of the IL33/IL1RL1 genes in asthma is biologically plausible.
Previous research has shown that the expression of IL-33 is involved in airway hyper-responsiveness, host defence, immune regulation, inflammation, serum IgE elevation and increased eosinophil counts [Citation6,Citation11]. In mouse models, IL-33 plays essential roles in the exacerbation of IgE-mediated airway inflammation and remodelling [Citation12]. However, whether serum levels of IL-33 correlate with asthma is currently unknown, which motivated part of the present study.
Genetic variation in both IL33 and IL1RL1 is strongly associated with asthma in genome-wide association studies (GWAS) in Mexican, European, North American and Brazilian populations. Wu et al. [Citation13] suggest that IL1RL1 single-nucleotide polymorphisms (SNPs) might contribute to Mexico childhood's asthma susceptibility. Moffatt et al. [Citation14] observed associations between asthma and SNPs linked to IL1RL1/IL18R1 and IL33 in a European population. IL-33 and IL1RL1 loci are associated with asthma risk in three ethnic groups of North American populations [Citation15]. Faruque et al. [Citation16] has replicated the association of IL33 upstream polymorphisms with asthma, previously identified in a predominantly European population, in an independent African-American population. IL33 and IL1RL1 variants have been associated with asthma in a Brazilian population [Citation17]. Grotenboer et al. [Citation18] found evidence that genetic variation at the IL33 and IL1RL1 loci translates into increased susceptibility for asthma. IL33, IL1R1 and RAD50 genes are also associated with the risk of asthma in a Chinese population [Citation19].
Therefore, in this study, our aim was to investigate whether SNPs of IL-33/IL1RL1 gene (rs16924159, rs1420102, rs1946131 and rs13431828) might have an effect on the susceptibility to asthma and the serum levels of IL-33 associated with asthma. Furthermore, we tried to assess the correlations between the IL-33 gene SNPs polymorphisms and the serum levels of IL-33 in the population of Xinjiang (China).
Subjects and methods
Study population
This case-control study included 556 unrelated participants: 201 adults and 355 children. The case groups comprised 279 patients with asthma (male 50.2%, female 49.8%). The diagnosis of asthma was according to the criteria of the Global Initiative for Asthma guidelines [Citation20]. The control groups included 277 healthy people (male 53.4%, female 46.6%). The study design and the patient data collection were conducted with the approval of the Ethics Committee of The Affiliated Hospital of Shihezi College (Xinjiang, China). The participants or their legal guardians signed informed consent forms.
Enzyme-linked immunosorbent assay (ELISA)
The levels of IL-33 were measured by ELISA according to the manufacturer's instructions (Uscn Life Science, Inc. Number: SEB980Hu).
Genotyping analysis
Genomic DNA was isolated from peripheral venous blood, using a genomic DNA extraction kit (NO.DP318, TIANGEN, Biotech, Beijing, China) according to the manufacturer's instructions. The concentration and purity of the isolated genomic DNA was checked by Nanodrop2000; its quality and integrity were analyzed using agarose gel electrophoresis. The DNA samples were then stored at −80 °C.
Four SNPs were chosen from the accessible literature, the National Center for Biotechnology Information (NCBI) database, HapMap and 1000Genomes. The selection criteria were as follows: functionality, ability to tag the haplotypes spanning the regions, previous clinical associations with asthma and high allele frequency (MAF >0.03). The genotyping of all 556 participants was performed using the MassARRAY® MALDI-TOF SNP genotyping system (Sequenom Inc., San Diego, Calif., USA). Polymerase chain reaction (PCR) amplification (GeneAmp PCR System 9700, Applied Biosystems Inc) was conducted for the loci carrying the examined SNPs. All of them were normal and showed successful amplification. Primer Premier 5.0 Software was used for site-specific primer set selection. Any deviations of the genotypes were excluded from further analyses. The primers used in this study are summarised in .
Table 1. PCR primers used in this study.
Table 2. Demographics of patients with asthma and controls.
Statistical analysis
The t-test was used to compare continuous variables and the Chi-square test for categorical variables between patients with asthma and controls. Measurements with a normal distribution of the data are expressed as mean values with standard deviation (x ± s) or as n (%), unless stated otherwise. The IL-33 serum level and asthmatic characteristics differences between genotypes were tested by the Kruskal–Wallis test (three groups) or t-test (two groups). The allele frequency and deviation of the genotype, which was estimated using the Hardy–Weinberg equilibrium (HWE), was assessed by the Chi-square test. Odds ratios (ORs) and p-values were calculated by multivariate unconditional logistic regression and p-values of less than 0.05 were considered significant. Data analysis was performed using the Statistical Package for Social Sciences (SPSS) 22.0 (IBM, NY, USA) and Graph Pad Prism 6.0 (Graph Pad Software, Inc., San Diego, CA, USA).
Results and discussion
The demographic characteristics of the study subjects are summarized in . The case and control groups were matched with respect to age and sex.
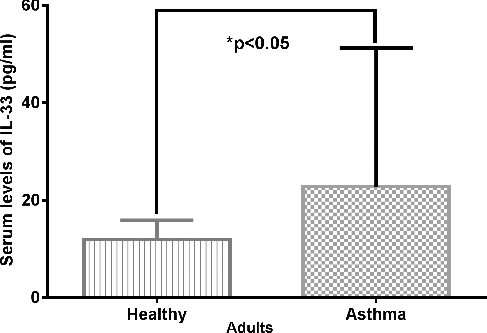
The IL-33 levels were measured in 279 patients with asthma and 277 control subjects. Most of the serum IL-33 levels in children were below the limits of detection. The undetectable serum levels of IL-33 in children might be due to mild asthmatic bronchitis, hemolytic, lipemic and precipitation samples. The mean serum IL-33 level of adults with asthma was 22.80 ± 28.49 pg/mL, whereas in the control group, it was 11.95 ± 3.92 pg/mL. The patients with asthma had a significantly higher level of serum IL-33 than the controls (): 22.80 ± 28.49 pg/mL vs. 11.95 ± 3.92 pg/mL (p < 0.05). These results are in accordance with previous evidence that there could be a positive correlation between the serum level of IL-33 and asthma. IL-33 expression is increased in the airway smooth muscle cells in asthma patients compared with controls, proposing IL-33 as a novel inflammatory marker of severe and refractory asthma [Citation21]. Gluck et al. [Citation22] have reported higher levels of IL-33 in the serum of patients with intermittent allergic rhinitis than in bronchial asthma/controls, and correlation between the serum level of IL-33 and the disease severity. Hamzaoui et al. [Citation23] have observed that the values of soluble ST2 and IL-33 in induced sputum correlate with the disease activity in young asthmatic children. Given these results and our data, the circulating levels of IL-33 may be a characteristic of airway inflammation and could be considered as a marker of asthma severity.
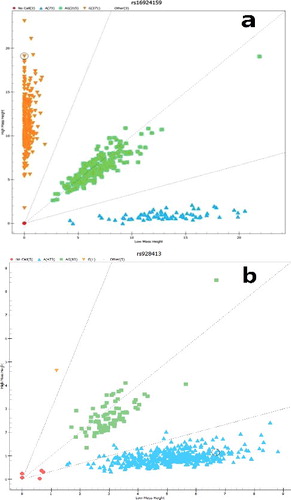
The characteristics of the four SNPs (including their position, genotype frequency and MAF) used in the study are shown in . The frequencies of alleles were in accordance with the Hardy–Weinberg equilibrium in each group. A representative mass spectrum scatterplot classification diagram of two genotypes is shown in . The genotype and allele frequencies of the four SNPs (i.e. rs16924159 and rs928413 in IL-33, and rs1420102 and rs13431828 in IL1RL1) are summarized in . The genotyping success rate was 97.8%–100%. The frequencies of the GG, AG and AA alleles of rs16924159 were 50.5, 32.3 and 17.2% in the cases and 46.6, 45.1 and 8.3% in the controls, respectively. There was a dramatic difference in the distribution of genotypes between the cases and the controls (OR = 0.659, 95% CI: 0.459–0.945; OR = 1.909, 95% CI: 1.100–3.314). However, the G allele of rs16924159 had no significant association with risk of asthma compared with the A allele (OR = 1.120, 95% CI: 0.870–1.441). The frequencies of the CC and TT alleles of rs1420102 were 26.5 and 27.2% in asthma patients and 32.9 and 19.1%, respectively, in controls. The T and TT genotypes were associated with a significant increased risk of asthma compared with the C and CC genotypes (OR = 1.337, 95% CI: 1.056–1.693, OR = 1.763, 95% CI: 1.107–2.810). However, the rs928413A/G genotypes, the rs13431828T allele and TT genotypes were not significantly associated with risk of asthma.
Table 3. Main characteristics of the four SNPs in the study.
Table 4. Genotype and allele frequency distributions of rs16924159, rs1420102, rs1946131and rs13431828 in patients with asthma and controls.
The association between IL33SNPs and allergic asthma or asthma has been reported previously [Citation24–27]. The IL-33 rs1412426 was firstly reported in a GWAS study, showing evidence that the T allele is associated with the development of atopic asthma in Europeans [Citation28]. Another GWAS identified IL-33 rs928413 as a susceptibility locus for childhood asthma [Citation29]. Chen et al. [Citation19] also found that allele G of rs928413 confers a remarkably higher risk of asthma. Grotenboer et al. [Citation18] describes the rs16924159 genetic variation at the IL33 locus which translates into increased susceptibility to asthma. Consistent with these findings, our results suggest that each one alone, i.e. rs16924159AA or rs1420102TT, is a variant for susceptibility to development of asthma.
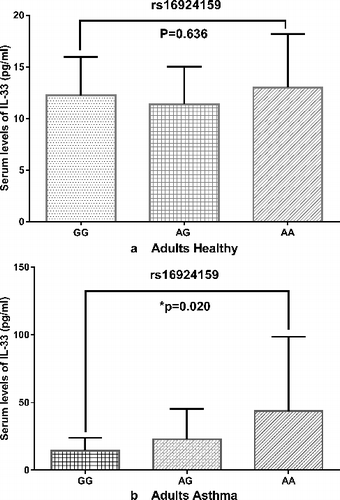
Generally, the serum IL-33 levels were higher in the patients with asthma (22.80 ± 28.49 pg/mL) than in the control subjects (11.95 ± 3.92 pg/mL) (p < 0.05) (). In a separate experiment, the effect of the IL33 rs16924159 polymorphisms on the IL-33 level was determined in adults with asthma. The patients carrying the rs16924159AA genotype presented with higher IL-33 levels (43.61 ± 55.07 pg/mL) than the AG (22.94 ± 22.51 pg/mL) and GG group (14.55 ± 9.43 pg/mL) ((a)). However, the rs928413AA/GG variation and the IL-33 levels in the control subjects showed no significant differences. The rs7044343 CC genotype of the IL-33 gene has been associated with decreased risk of developing rheumatoid arthritis and with low serum IL-33 [Citation30]. Monocytes from individuals with the rs7044343 CC genotype produce higher levels of IL-33 (32.08 ± 24.30 pg/mL) than those from patients with CT (16.32 ± 6.23) (P = 0.005) and TT (17.10 ± 5.48 pg/mL) (P = 0.007) genotypes [Citation31]. The present study revealed an association between the genotype frequency of rs16924159 and IL-33 levels. The subjects who carry the AA allele for rs16924159 had higher IL-33 levels than the patients with the GG and AG alleles. This result suggested that the increase in the IL-33 levels in patients with asthma may be controlled by the rs16924159polymorphism.
Figure 3. Serum levels of IL-33 with three genotypes of rs16924159 in healthy controls (a) and asthmatic patients (b).
IL-33 binds to IL-1 receptor-like 1 (IL1RL1-b) and forms a receptor complex with IL-1 receptor-associated protein (IL1RAcP). This receptor complex induces release of allergic and eosinophilic mediators, such as IL-5 and IL-13, resulting in eosinophilic inflammation [Citation5,Citation18,Citation32,Citation33]. Polymorphisms (rs13431828, rs1420102 and rs928413) of IL1RL1 associated with the risk of asthma have been identified in different ethnic groups [Citation13,Citation19,Citation28,Citation33]. However, rs13431828 and rs928413 had no significant association with risk of asthma in the population of Xinjiang.
Several limitations, however, need to be acknowledged. We were not able to detect the serum levels of IL-33 in many of the study participants. When interpreting the results from this study, we could not exclude potential effects of environmental risk factors, and there might have been undiagnosed asthmatics among the controls. Within these limitations, our research supported the conclusion that there are differences in these SNPs and the risk of asthma in different populations with distinct genetic backgrounds and ethnicities. ethnic differences.
Conclusions
In our study involving 279 Xinjiang Chinese patients with asthma, we demonstrated that the TT genotype of rs1420102 and the AA genotype of rs16924159 contribute to increased asthma susceptibility, while the TT genotype of rs13431828 does not contribute to asthma susceptibility. In addition, within the limitations of this study, the results provided evidence that expression of serum IL-33 was associated with asthma in participants; rs16924159 AA was associated with higher IL-33 levels and the rs16924159 G-A variant may affect the expression of IL-33. Further studies are needed to investigate the causal link between the IL33-IL1RL1 pathway genotype polymorphisms and IL-33 levels.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Martinez FD, Vercelli D. Asthma. Lancet. 2013;382(9901):1360–1372.
- Croisant S. Epidemiology of asthma: Prevalence and burden of disease. Adv Exp Med Biol. 2014;795:17–29.
- Holgate ST. Genetic and environmental interaction in allergy and asthma. J Allergy Clin Immunol. 2000;104(6):1139–1146.
- Mcleish S, Turner WS. Gene-environment interactions in asthma. Arch Dis Childhood. 2007;92(11):1032–1105.
- Liew FY, Pitman NI, Mcinnes IB. Disease-associated functions of IL-33: the new kid in the IL-1 family. Nat Rev Immunol. 2010;10(2):103–110.
- Schmitz J, Owyang A, Oldham E, et al. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity. 2005;23(5):479–490.
- Smithgall MD, Comeau MR, Yoon BRP, et al. IL-33 amplifies both Th1- and Th2-type responses through its activity on human basophils, allergen-reactive Th2 cells, iNKT and NK cells. Int Immunol. 2008;20(20):1019–1030.
- Chackerian AA, Oldham ER, Murphy EE, et al. IL-1 receptor accessory protein and ST2 comprise the IL-33 receptor complex. J Immunol. 2007;179(4):2551–2555.
- Moussion C, Ortega N, Girard JP. The IL-1-like cytokine IL-33 is constitutively expressed in the nucleus of endothelial cells and epithelial cells in vivo: a novel ‘alarmin’ ? Plos One. 2008 [cited 2017 Dec 15];3(10):e3331. DOI:10.1371/journal.pone.0003331.
- Savenije OE, Mahachie John JM, Granell R, et al. Association of IL33-IL-1 receptor-like 1 (IL1RL1) pathway polymorphisms with wheezing phenotypes and asthma in childhood. J Allergy Clin Immunol. 2014;134(1):170–177.
- Kearley J, Buckland KF, Mathie SA, et al. Resolution of allergic inflammation and airway hyperreactivity is dependent upon disruption of the T1/ST2-IL-33 pathway. Am J Respir Crit Care Med. 2009;179(9):772–781.
- Mizutani N, Nabe T, Yoshino S. Interleukin-33 and alveolar macrophages contribute to the mechanisms underlying the exacerbation of IgE-mediated airway inflammation and remodelling in mice. Immunology. 2013;139(2):205–218.
- Wu H, Romieu I, Shi M, et al. Evaluation of candidate genes in a genome-wide association study of childhood asthma in Mexicans. J Allergy Clin Immunol. 2010;125(2):658–661.
- Moffatt MF, Gut IG, Demenais F, et al. A large-scale, consortium-based genome-wide association study of asthma. N Engl Med. 2010;363(13):1211–1221.
- Torgerson DG, Ampleford EJ, Chiu GY, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43(9):887–892.
- Faruque MU, Chen G, Doumatey AP, et al. Transferability of genome-wide associated loci for asthma in African Americans. J Asthma. 2016;54(1):1–8.
- Queiroz GA, Costa RS, Alcantaraneves NM, et al. IL33 and IL1RL1 variants are associated with asthma and atopy in a Brazilian population. Int J Immunogenet. 2017;44(2):51–61.
- Grotenboer NS, Ketelaar ME, Koppelman GH, et al. Decoding asthma: translating genetic variation in IL33 and IL1RL1 into disease pathophysiology. J Allergy Clin Immunol. 2013;131(3):856–865.
- Chen J, Zhang J, Hu H, et al. Polymorphisms of RAD50, IL33 and IL1RL1 are associated with atopic asthma in Chinese population. Tissue Antigens. 2015;86(6):443–447.
- Bateman ED. Global strategy for asthma management and prevention: GINA executive summary. Eur Respir. 2008;31(1):143–178.
- Préfontaine D, Lajoiekadoch S, Foley S, et al. Increased expression of IL-33 in severe asthma: evidence of expression by airway smooth muscle cells. J Immunol. 2005;175(4):2702–2708.
- Gluck J, Rymarczyk B, Rogala B. Serum IL-33 but not ST2 level is elevated in intermittent allergic rhinitis and is a marker of the disease severity. Inflamm Res. 2012;61:547–550.
- Hamzaoui A, Berraies A, Kaabachi W, et al. Induced sputum levels of IL-33 and soluble ST2 in young asthmatic children. J Asthma. 2013;50(8):803–809.
- Reijmerink NE, Postma DS, Bruinenberg M, et al. Association of IL1RL1, IL18R1,and IL18RAP, gene cluster polymorphisms with asthma and atopy. J Allergy Clin Immunol. 2008;122(3):651–654.e8.
- Reijmerink NE, Bottema RWB, Kerkhof M, et al. TLR-related pathway analysis: novel gene–gene interactions in the development of asthma and atopy. Allergy. 2010;65(2):199–207.
- Ferreira MA, Mcrae AF, Medland SE, et al. Association between ORMDL3, IL1RL1 and a deletion on chromosome 17q21 with asthma risk in Australia. Eur J Hum Genet. 2011;19(4):458–464.
- Belpinati F, Malerba G, Trabetti E, et al. Association of childhood allergic asthma with markers flanking the IL33 gene in Italian families. J Allergy Clin Immunol. 2011;128(3):667–668.
- Gudbjartsson DF, Bjornsdottir US, Halapi E, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41(3):342–347.
- Bønnelykke K, Sleiman P, Nielsen K, et al. A genome-wide association study identifies CDHR3 as a susceptibility locus for early childhood asthma with severe exacerbations. Nat Genet. 2014;46(1):51–55.
- Li C, Mu R, Guo J, et al. Genetic variant in IL33 is associated with susceptibility to rheumatoid arthritis. Arthritis Res Ther. 2014 [cited 2017 Dec 15];16(2):R105. DOI:10.1186/ar4554.
- Angelesmartínez J, Posadassánchez R, Llorente L, et al. The rs7044343 polymorphism of the interleukin 33 gene is associated with decreased risk of developing premature coronary artery disease and central obesity, and could be involved in regulating the production of IL-33. Plos One. 2017 [cited 2017 Dec 15];12(1):e0168828. DOI:10.1371/journal.pone.0168828.
- Kakkar R, Lee RT. The IL-33/ST2 pathway: therapeutic target and novel biomarker. Nat Rev Drug Discov. 2008;7(10):827–840.
- Savenije OEM, Kerkhof M, Reijmerink NE, et al. Interleukin-1 receptor–like 1 polymorphisms are associated with serum IL1RL1-a, eosinophils, and asthma in childhood. J Allergy Clin Immunol. 2011;127(3):750–756.