Abstract
The aim of this study was to investigate the application of amniotic fluid cell lines with chromosomal abnormalities in the external quality assessment (EQA) of prenatal karyotype analysis. Simian virus 40 large T antigen (SV40LT) gene was transfected into amniotic fluid cells with 46,XY,t(15;17)(q11;q11) and 46,XY by liposome transfection. Control cell lines were produced by mixing the above two in ratios of 1:19, 1:1 and 9:1, respectively. The cells were then frozen at −30 °C, mailed between laboratories, prepared for chromosome analysis, photographed with karyotype, and evaluated. After the two amniotic fluid cell lines and the control cell lines were serially passaged for 10 generations, they were still spindle-shaped with adherent growth. The 46,XY,t(15;17)(q11;q11) line preserved the original amniotic fluid karyotype, whereas a small number of cells with 46,XY, had changes of chromosomal structure. After mailing between laboratories, the abnormal karyotype was scored to constitute, 10.8%, 54.2% and 81.2% on average in the 1:19, 1:1 and 9:1 samples, respectively. There were significant differences (P < 0.05) between the actual scores and the theoretically expected ones with respect to the controls with 5% and 90% abnormal karyotype, whereas no significant difference (P > 0.05) with respect to the ratio of 50%. Overall, we demonstrated that the SV40LT gene could immortalize amniotic fluid cells with chromosomal abnormalities. The immortalized cells with and without chromosomal abnormalities mixtured in different ratios could then serve as control cells in the EQA of prenatal karyotype analysis.
Introduction
Chromosome karyotype analysis is the diagnosis of chromosomal disease by analyzing the number and structure of chromosomes. It has been widely used in prenatal diagnosis and its demand is increasing due to the development of non-invasive prenatal genetic testing and the increase of pregnant women in advanced maternal age [Citation1]. Chromosome karyotype analysis is the gold standard for the diagnosis of chromosomal disease, and its accuracy is largely influenced by cell culture, chromosome preparation, band identification and diagnostic techniques. Therefore, it is important to pay attention to quality control [Citation2,Citation3]. Sikkema-Raddatz et al. [Citation4] previously studied the quality control of prenatal cytogenetic diagnosis; however, they only focused on the influencing factors on pre-culturing specimens of amniotic fluid cells and chorionic villi cells. Lymphocytes with chromosome abnormalities as the quality control cells are used in the external quality assessment (EQA) of chromosome karyotype analysis, but lymphocytes and the amniotic fluid cells used in prenatal karyotype analysis are not homologous cells [Citation5]. Other reports have proposed the application of karyotype images of chromosomally abnormal cells as quality control [Citation6,Citation7]. Although this method is simple and has been implemented, it could not evaluate the details of cell culture, chromosome preparation and G-banding [Citation3]. However, standardized diagnostic protocols are also important for the quality control system [Citation8–10]. Thus, ISO15189 asks to estimate the measurement uncertainty, which is calculated using a combination of both the long-term imprecision internal quality control results and bias, on the basis of EQA results [Citation11]. So, quality control materials should be amplified on the premise of maintaining the same characteristics and should reflect the experimental process. To this end, in this study, the interventricular mass evaluation method for karyotype analysis of amniotic fluid cells was studied with chromosomal abnormal amniotic fluid cell lines as a quality control sample.
Materials and methods
Construction of amniotic fluid cell lines
Amniotic fluid cell lines were constructed similar to the methods described previously [Citation12–15]. Briefly, Simian virus 40 large T antigen (SV40LT) recombinant vector SV40LTag-pcDNA3.1) (-) was constructed and transfected into 46,XY,t(15;17)(q11;q11) and 46,XY primary amniotic fluid cells by liposome transfection. Then the cells were screened by G418 and the positive cell clones were passaged repeatedly. Well-grown cells of 3–5 generations each were harvested and frozen at −196 °C in liquid nitrogen.
Preparation of control cell lines
The amniotic fluid cells with karyotype 46,XY,t(15;17)(q11;q11) and 46,XY stored in liquid nitrogen were thawed at 37 °C, subcultured for 3–5 generations and were harvested. Then the cells were suspended in phosphate buffered saline and were counted with a hemocytometre. Cell suspensions were prepared at a final concentration of 5 × 105/mL and the two kinds of cells were accurately mixed at ratios of 1:19, 1:1 and 9:1 to obtain samples with 5%, 50% and 90% of the karyotype 46,XY,t(15;17)(q11;q11), respectively. After passaging for another 3–5 generations, the cells were treated with a hypotonic solution and were fixed according to conventional chromosome preparation methods. Finally, the cells were divided and stored at −30 °C for future use.
Fluorescence in situ hybridization (FISH) [Citation16] assay
FISH assay employed the AneuVysion Assay Kit (Abbot Molecular, US) to detect 21, 18, 13, X and Y chromosomes of the control cell lines according to its manual and the images were captured with a BX50 Olympus microscope. FISH experiments were performed by the same technician.
Trial with control cell lines for karyotype analysis in EQA
A total of 90 tubes of the 5%, 50% and 90% control cells, that is, 30 tubes each, kept frozen in a refrigerator at −30 °C for 3–6 months were taken. The tubes were packed together into an extraction carton and sent back and forth between different laboratories. The mailing time was 48–72 hours and the temperature was 15 ∼ 25 °C. After the recovery, cells were prepared for chromosome GTG-banding [Citation17]. Then, a total of nine samples, including three copies of each ratio of control cells, were distributed to 10 technical staff members for conventional chromosome karyotype analysis.
Data analysis
The data are presented as mean values. Statistical analysis was performed using Chi-Squared Test in R 3.3.1 software (https://www.r-project.org/).
Results and discussion
Amniotic cell line construction
With the extensive use of chromosome karyotype analysis in prenatal diagnosis, great attention has been paid to quality control [Citation3–7]. Unfortunately, due to the lack of birth defects resources and homologous cells, which are the necessary quality control materials, it is difficult to carry out internal quality control and EQA of karyotype analysis [Citation3]. So far, only a few cell lines could be used for quality control, including lymphocytes and primary cells remaining after prenatal diagnosis or viable subcultured cells staying adhered to the culture flask after the passage, with uncommon and complicated chromosomal abnormalities [Citation5,Citation18]. However, lymphocytes are not homologous to amniotic cells [Citation5]. Since primary cells are not immortalized, they are difficult to grow in large quantities [Citation18].
The Simian virus 40 large T antigen (SV40LT) gene can result in the immortalization of most human cells [Citation15]. Immortalized cells could be cultured for more than 350 generations in vitro, and retain the differentiation phenotype and biological characteristics of their original cells [Citation13,Citation19]. In particular, human gastric mucosa, nasopharyngeal, tracheal epithelial cells, skin and embryonic lung fibroblasts, which are all surface cells homologous to the original amniotic fluid cells, could be immortalized based on SV40LT gene transfection [Citation12,Citation13,Citation20–22]. Therefore, we chose SV40LT for transfecting cells with karyotype 46,XY,t(15;17)(q11;q11) and 46,XY and passaged for amplification and EQA in karyotype analysis.
In this study, two amniotic cell lines, 46,XY,t(15;17)(q11;q11) and 46,XY, were constructed, respectively. They were passaged to the 10th generation and were still spindle-shaped, with adherent growth in a monolayer (). We found that the 46,XY,t(15;17)(q11;q11) cells preserved the original amniotic fluid karyotype (). However, a small number of cells with 46,XY, had chromosomal structural changes. For example, one of the long arms of chromosome 11 increased by 1 deep band [der (11)], which increased the difficulty of karyotype analysis (). These results demonstrated that SV40LT could immortalize amniotic cell lines with abnormal karyotype. Additionally, they could be then grown, thus solving the problem of large-scale cultivation of quality control cells, and used to evaluate cell culture and chromosome preparation in details.
Figure 1. Spindle-shaped and adherent amniotic fluid cells passaged for 10 generations after transfection with SV40LT. Cells with 46,XY, t(15;17)(q11;q11) karyotype (A) and 46,XY karyotype (B).
Note: Images captured under a phase contrast microscope (200X).
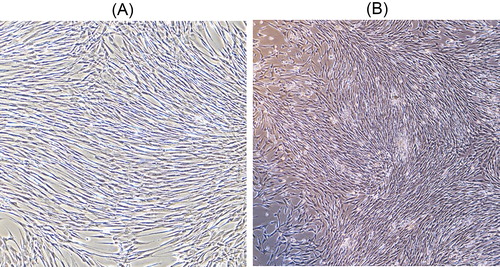
Figure 2. Representative images from chromosome karyotype analysis of the 10th generation of cells with 46,XY,t(15;17)(q11;q11) karyotype (A) and 46,XY with a changed chromosomal karyotype (B).
Note: (A) The arrows point at the abnormal chromosome. (B) The arrow indicates that one of the long arms of chromosome 11 increased by 1 deep band [der (11)].
![Figure 2. Representative images from chromosome karyotype analysis of the 10th generation of cells with 46,XY,t(15;17)(q11;q11) karyotype (A) and 46,XY with a changed chromosomal karyotype (B).Note: (A) The arrows point at the abnormal chromosome. (B) The arrow indicates that one of the long arms of chromosome 11 increased by 1 deep band [der (11)].](/cms/asset/082835a7-d796-45d9-b15a-fc14cb32d169/tbeq_a_1530072_f0002_c.jpg)
FISH
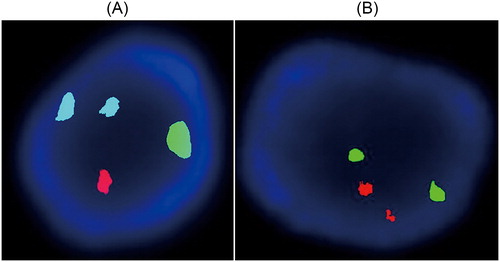
To prepare control cell lines, the two different cell lines were mixed in different ratios. The control cells were prepared by mixing 46,XY,t(15;17)(q11;q11) and 46,XY according to the quality control requirements. In order to further validate the karyotype of the control cell lines, FISH experiments were performed for 21, 18, 13, X and Y chromosome detection. The results showed a clear fluorescence signal. There were two blue, one green, and one red fluorescent signal, representing two 18 chromosomes, one X, and one Y chromosome, respectively (). As illustrated in , each had two green and red fluorescent signals, indicating two 13 and 21 chromosomes.
Figure 3. FISH analysis of fetal amniotic cells with 46,XY,t(15;17)(q11;q11) karyotype. (A) Two blue, one green and one red fluorescent signal, representing two 18 chromosomes, one X and one Y chromosome, respectively. (B) Two green and two red fluorescent signals, respectively, representing two 13 and two 21 chromosomes.
EQA results for control cell lines
In order to determine if the control cell lines would be suitable for karyotype analysis, EQA was undertaken. After a total of 90 samples of quality control cell lines were mailed between labs, EQA test by 10 technicians showed that the quality control cell lines with 5%, 50% and 90% abnormal cells got results returned for 24, 26 and 25 samples, the rates of return being 80.0%, 86.7% and 83.3%, respectively. The average reported abnormal karyotype rates were 10.8%, 54.2% and 81.2%, respectively (). For the control samples containing 5% and 90% cells with abnormal karyotype, there were significant differences (P < 0.05) between the theoretically expected and the EQA results, whereas there was no significant difference (P > 0.05) with respect to the samples with a ratio of 50% (). These findings suggested the obtained amniotic fluid cell lines with chromosomal abnormalities could potentially be used in the EQA of prenatal karyotype analysis.
Table 1. EQA results for the control cell lines.
In theory, cells after passaging for 10 generations could meet the needs of laboratory batch use. In this study, we noted significant differences between the average determined value of abnormal karyotypes and that of the generated samples with abnormal karyotypes of 5% and 90%. This could be presumably due to paying overly special attention to the cells with chromosomal abnormalities when present in a low proportion, while more easily ignoring some of them when present in a high proportion. Additionally, there could be errors in the theoretical proportions vs. the actual proportions. There is also the possibility that the chromosome structure in a small number of cells may have changed in the course of repeated passage of the amniotic fluid cell lines. The variation in chromosome structure made it difficult to analyze the karyotype; therefore, it is necessary to carry out comprehensive analysis by combining the relevant diagnostic methods to confirm the quality control cells.
Conclusions
In this study, we tested the karyotypes in a simple control cell line and conducted preliminary tests with a limited number of technicians. Amniotic fluid cell lines with chromosomal abnormalities could solve the problem of the lack of ideal quality control materials in prenatal karyotype analysis. The proposed cell lines showed potential for future use in EQA. In addition, they could be applied to practical work in a standardized way. However, more works are needed to standardize EQA for karyotype analysis.
Funding
This study was Supported by the National Natural Science Foundation of China under grant number 81671467, the Medicine and Health Science and Technology Plan Projects in Zhejiang Province under grant number 201494417, the Opening Foundation of the State Key Laboratory for Diagnosis and Treatment of Infectious Diseases and Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, and The First Affiliated Hospital of Medical College, Zhejiang University under grant number 2017KF06.
Disclosure statement
The authors declare that they have no competing interests.
References
- Norton ME, Jelliffe-Pawlowski LL, Currier RJ. Chromosome abnormalities detected by current prenatal screening and noninvasive prenatal testing. Obstet Gynecol. 2014;124:979–986.
- Hastings RJ, Maher EJ, Quellhorst-Pawley B, et al. An Internet-based external quality assessment in cytogenetics that audits a laboratory's analytical and interpretative performance. Eur J Hum Genet. 2008;16:1217–1224.
- Wang W, Chen Y, Chen X, et al. Result survey analysis of prenatal chromosome karyotyping in an external quality assessment program. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2014;31:483–486.
- Sikkema-Raddatz B, Suijkerbuijk R, Bouman K, et al. Quality aspects of prenatal cytogenetic diagnosis: determining the effect of various factors involved in handling amniotic fluid and chorionic villus material for cytogenetic diagnosis. Prenat Diagn. 2006;26:791–800.
- Weng BH, Cai JP, Wang XM, et al. Establishment of lymphocyte cell lines with abnormal chromosome karyotypes and its application in external quality assesment for chromosome karyotype analysis. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2007;24:689–691.
- Weng B, Li X. An external quality assessment scheme for prenatal detection of rare chromosomal abnormalities. Clin Chim Acta. 2012;413:1721–1724.
- Weng BH, Lu YE, Li X. External quality assessment of prenatal diagnosis of a rare and subtle chromosomal structural abnormality. Eur Rev Med Pharmacol Sci. 2013;17:2605–2608.
- Hannema SE, de Rijke YB. Improving laboratory assessment in disorders of sex development through a multidisciplinary network. Sex Dev. 2018;12:135–139.
- Delis H, Christaki K, Healy B, et al. Moving beyond quality control in diagnostic radiology and the role of the clinically qualified medical physicist. Phys Med. 2017;41:104–108.
- Driscoll AJ, Karron RA, Morpeth SC, et al. Standardization of laboratory methods for the PERCH Study. Clin Infect Dis. 2017;64:S245–S252.
- Padoan A, Antonelli G, Aita A, et al. An approach for estimating measurement uncertainty in medical laboratories using data from long-term quality control and external quality assessment schemes. Clin Chem Lab Med. 2017;55:1696–1701.
- Scholte BJ, Kansen M, Hoogeveen AT, et al. Immortalization of nasal polyp epithelial cells from cystic fibrosis patients. Exp Cell Res. 1989;182:559–571.
- Reddel RR, Ke Y, Gerwin BI, et al. Transformation of human bronchial epithelial cells by infection with SV40 or adenovirus-12 SV40 hybrid virus, or transfection via strontium phosphate coprecipitation with a plasmid containing SV40 early region genes. Cancer Res. 1988;48:1904–1909.
- Chang PL, Gunby JL, Tomkins DJ, et al. Transformation of human cultured fibroblasts with plasmids carrying dominant selection markers and immortalizing potential. Exp Cell Res. 1986;167:407–416.
- Kirchhoff C, Araki Y, Huhtaniemi I, et al. Immortalization by large T-antigen of the adult epididymal duct epithelium. Mol Cell Endocrinol. 2004;216:83–94.
- Basel-Vanagaite L, Zevit N, Har Zahav A, et al. Transient infantile hypertriglyceridemia, fatty liver, and hepatic fibrosis caused by mutated GPD1, encoding glycerol-3-phosphate dehydrogenase 1. Am J Hum Genet. 2012;90:49–60.
- Simons A, Shaffer LG, Hastings RJ. Cytogenetic nomenclature: Changes in the ISCN 2013 Compared to the 2009 Edition. Cytogenet Genome Res. 2013;141:1–6.
- Yu X, Weng B, Li H, et al. Preparation and application of cells used in the external quality assessment of karyotype analysis. Biomed Res. 2017;28:5074–5077.
- Campisi J. Replicative senescence: an old lives' tale? Cell. 1996;84:497–500.
- Kim SH, Banga S, Jha KK, et al. SV40-mediated transformation and immortalization of human cells. Dev Biol Stand. 1998;94:297–302.
- Ozer HL. SV40-mediated immortalization. Prog Mol Subcell Biol. 2000;24:121–153.
- Tevethia MJ, Ozer HL. SV40-mediated immortalization. Methods Mol Biol. 2001;165:185–199.
