Abstract
Although stresses induce generation of reactive oxygen species (ROS), which are highly reactive and toxic, and cause severe damage to cellular components; plants have very efficient enzymatic ROS-scavenging mechanisms. Despite the substantial knowledge produced about these enzymes, we still have limited knowledge regarding their expression patterns in relation to the stress type, duration and strength. Thus, taking advantage of microarray data, this work evaluated the abiotic stresses (salt, cold, heat and light) induced regulation of six antioxidant enzymes, superoxide dismutase (SOD), catalase (CAT), ascorbate peroxidase (APX), glutathione peroxidase (GPX), monodehydroascorbate reductase (MDHAR) and dehydroascorbate reductase (DHAR), in 10 natural Arabidopsis ecotypes. The expression profiles of 36 genes encoding six enzymatic antioxidants including CSD1-3, FSD1-3, MSD1-2, CAT1-3, APX1-6, APXT, APXS, GPX1-8, MDAR1-5 and DHAR1-4 were investigated. In particular, FSD1, FSD2, CSD1 and CSD2 genes coding for SOD; CAT2 and CAT3 for CAT; APX3-6, APXT and APXS for APX; GPX1, GPX2, GPX5, GPX6 and GPX7 for GPX; MDAR2-4 for MDHAR; and DHAR1 and DHAR3 for DHAR families appeared to be more differentially expressed under given stress conditions. Primarily, high light as well as salt and cold stresses considerably up-regulated the gene expression, whereas cold stress significantly led to the down-regulation of genes. The overall expression pattern of ecotypes suggested that the studied Arabidopsis genotypes had different stress tolerance.
Introduction
Abiotic stresses induce the overproduction of reactive oxygen species (ROS) which are highly reactive and toxic, and cause severe damage to cellular molecules [Citation1,Citation2]. ROS induce damage (1) via peroxidation of lipids, which increases the membrane leakiness, reduces the membrane fluidity and damages the membrane proteins, ion channels, enzymes and receptors [Citation3–5]; (2) through oxidation of proteins, mostly irreversible, which hinders or changes the protein activities and makes proteins more susceptible to proteolytic attack [Citation6] and (3) by DNA damage with base deletions and modifications, strand breaks, cross-links and pyrimidine dimers [Citation7], resulting in decreased or damaged protein syntheses, cell membrane destruction, replication errors, genomic instability and transcription arrest or induction [Citation8,Citation9]. ROS could be either free radicals such as OH· (hydroxyl radical), O2.−(superoxide radical), HO2· (perhydroxy radical) and RO· (alkoxy radical) or non-radical forms such as H2O2 (hydrogen peroxide) and 1O2 (singlet oxygen) [Citation1,Citation10–12]. They are mainly generated in mitochondria, chloroplasts and peroxisomes as byproducts of various metabolic pathways resulting from intense electron flow or highly oxidizing activities [Citation13,Citation14]. Besides, cytoplasm, endoplasmic reticulum (ER) and apoplast are also other substantial ROS production sources in plants [Citation15]. ROS generation or accumulation is thus an inevitable outcome of normal metabolic processes in plants [Citation1]. However, in steady state conditions, there is an equilibrium between ROS production and its scavenging; any disturbance of this equilibrium by biotic/abiotic stress factors could increase the intracellular ROS levels [Citation16,Citation17]. Moreover, ROS like H2O2 play dual roles in plants regulating many crucial metabolic processes, including acclimatory signalling [Citation18], growth and development [Citation19], cell cycle [Citation20], photosynthesis and photorespiration [Citation21], stomatal movement [Citation22] and senescence [Citation2]. Therefore, plants have very efficient ROS-scavenging mechanisms employing enzymatic and non-enzymatic components. For example, ascorbate peroxidase (APX; EC 1.11.1.11), glutathione peroxidase (GPX; EC 1.11.1.9), catalase (CAT; EC 1.11.1.6), glutathione reductase (GR; EC 1.6.4.2), superoxide dismutase (SOD; EC 1.15.1.1), dehydroascorbate reductase (DHAR; EC 1.8.5.1), monodehydroascorbate reductase (MDHAR; EC 1.6.5.4), glutathione-S-transferase (GST; EC 2.5.1.18) and guaicol peroxidase (GOPX; EC 1.11.1.7) as enzymatic machineries, and phenolic compounds, carotenoids, flavonoids, glutathione (GSH), alkaloids, ascorbic acid (AA), α-tocopherol and some amino acids as non-enzymatic defence molecules [Citation1,Citation23]. SOD is the most effective enzymatic antioxidant catalyzing the removal of O2.− with its dismutation, one O2.− being oxidized to O2 while another reduced to H2O2. There are three known SOD types based on their cofactors: Fe-SOD, Mn-SOD and Cu/Zn-SOD [Citation24]. The Arabidopsis genome was reported to contain three Cu/ZnSOD and three FeSOD genes, and one MnSOD gene [Citation25]. CAT has one of the highest turnover rates among all enzymes, removing H2O2 by directly converting it into H2O and O2. Its isoforms, including two in barley [Citation26]; three in Arabidopsis [Citation27], maize [Citation28], pumpkin [Citation29] and rice [Citation30]; and four in sunflower [Citation31] have been previously reported. APX functions in the scavenging of H2O2 in ascorbate-glutathione (ASH-GSH) and water-water cycles using ASH as the electron donor. Studies have reported eight APX genes in Arabidopsis, eight in rice, and seven in tomato [Citation32–34]. GPX is another important enzyme involved in the reduction of H2O2 and some other peroxides using GSH. Arabidopsis genome were reported to include eight GPX genes [Citation35,Citation36]. MDHAR is a FAD-enzyme catalyzing the reduction of monodehydroascorbate (MDHA) radicals in the presence of NAD(P)H. MDHAR cDNA has been cloned from a number of plant species, including Arabidopsis, cucumber, soybean, potato, tomato, pea, rice and leaf mustard [Citation37]. DHAR catalyzes the re-reduction of DHA (dehydroascorbic acid) to ascorbate by glutathione (GSH). The crystal structure of DHARs from Arabidopsis and their pivotal role of DHARs under photooxidative stress has been investigated [Citation38,Citation39]. GR is an important enzyme in the ASH-GSH cycle, playing an essential role in ROS defence by maintaining the reduced GSH level via catalyzing the NADPH dependent reaction of glutathione disulphide (GSSG). In a recent genome-wide study, two and three GR genes in Arabidopsis and rice (Oryza sativa) genomes, respectively, were reported [Citation40]. Considering the presence or abundance of these antioxidant enzymes in almost all plant cells in response to various stress conditions, it is widely acknowledged that ROS-scavenging is significant for plant survivability as well as for crop productivity [Citation24,Citation41,Citation42]. Although a great amount of research has produced substantial knowledge about these enzymes, we still seem to possess just a general idea of their complex roles under various conditions. Thus, taking advantage of microarray data, we investigated the abiotic stress-induced regulation of antioxidant enzyme genes in 10 different Arabidopsis ecotypes, with an ultimate goal of understanding the expression profiles of these enzymes under various stress factors. In this study, (1) the regulation of six Arabidopsis antioxidant enzyme genes, SOD, CAT, APX, GPX, MDHAR and DHAR, were investigated under cold, heat, high light and salt stress; (2) the type of stress factor that leads to significant up/down regulation of these genes was identified; and (3) the association of the expression pattern in relation to different ecotypes was investigated.
Materials and methods
Microarray data
The microarray data for this study was retrieved from the National Center for Biotechnology Information (NCBI) GEO datasets repository (ncbi.nlm.nih.gov/geo/) with record GSE41935 [Citation43,Citation44]. Three-week-old plants from 10 natural Arabidopsis ecotypes, An-1 (Antwerpern/Belgium), Cvi (Cape Verdia Islands), Col-0 (Columbia/United States), C24 (Coimbra/Portugal), Eri (Erigsboda/Sweden), Kas-1 (Kashmir/India), Kond (Kondara/Tajikistan), Kyo-2 (Kyoto/Japan), Ler (Landsberg/Poland) and Sha (Shakdara/Tajikistan), were subjected to salinity (100 mmol/L NaCl), cold (10 °C), heat (38 °C) and light (800 μmol photons m−2s−1) stresses for 3 hours. RNA was extracted from leaf samples. GPL16226 [NimbleGen A. thaliana 4x72K Array (080306_AT6_expr)] platform was employed for the studies. Three replicates of treatment-vs-control samples were processed using NCBI’s GEO2R tool to identify the differentially expressed genes across experimental conditions. Heatmaps were generated using CIMminer tool (discover.nci.nih.gov/cimminer/home.do) [Citation45].
Results and discussion
Utilizing the microarray data with record GSE41935 in NCBI’s GEO datasets, the expression profiles of six Arabidopsis antioxidant enzymes, SOD, CAT, APX, GPX, MDHAR and DHAR, under four different stress conditions, cold, heat, high light and salt, were evaluated in 10 natural Arabidopsis ecotypes originating from different geographical locations. Initially, a detailed literature and database search was conducted to compile the Arabidopsis antioxidant enzyme genes. A total of 36 genes were identified for six different antioxidant enzymes, of which SODs included eight members (CSD1-3, FSD1-3 and MSD1-2), CATs included three ones (CAT1-3), APXs eight (APX1-6, APXT and APXS), GPXs eight (GPX1-8), MDHAR five (MDAR1-5) and DHAR four (DHAR1-4) members. Then, the expression profiles of each antioxidant enzyme under given abiotic stresses were individually evaluated in all Arabidopsis ecotypes ().
Table 1. List of major ROS-scavenging enzymes in Arabidopsis.
Superoxide dismutase (EC 1.15.1.1)
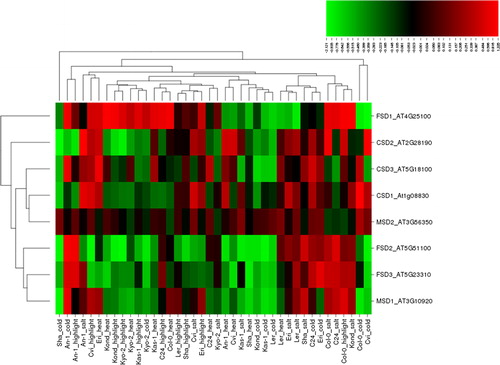
It is the most effective intracellular enzyme ubiquitously found in all aerobic organisms and in subcellular compartments exposed to ROS-derived oxidative stress [Citation1]. Many studies have reported significant changes in SOD activities under stress conditions of salt, cold, heat and high light exposure, which were investigated in this study. For example, salt stress treatment increased the SOD activity in tomato (Solanum lycopersicum) roots, chickpea (Cicer arietinum) and mulberry (Morus sp.) plants [Citation46–48]. Salt-treated leaf tissue extracts from chickpea exhibited significant SOD activity bands for MnSOD, FeSOD and Cu/ZnSOD [Citation49]. Salt and drought stresses upregulated the SOD activity in liquorice seedlings (Glycyrrhiza uralensis) and a MnSOD isoenzyme was identified in 2% NaCl exposed plants [Citation50]. Tea plants showed high SOD activity following 2 days of cold treatment, but decreased activity after 10 days [Citation51]. SOD activity remained high up to 4 days in cold treated chickpea plants but subsequently slightly declined [Citation52]. Two-day cold acclimation in potato (Solanum tuberosum) increased the SOD activity and conferred improved freezing tolerance [Citation53]. SOD activity along with the activities of other H2O2-scavenging enzymes were reported to be increased with light intensity, particularly in Mg-deficient leaves [Citation54]. Under high light, drought stress crucially increased the SOD activity in Picea asperata seedlings [Citation55]. The expression profiles of 18 cotton (Gossypium hirsutum) GhSOD genes were analyzed under different abiotic stresses and they showed ectopic expression and important roles in the protection against ROS [Citation56]. The antioxidant enzyme activities in three mulberry cultivars were found high under high temperature treatment [Citation57]. Moreover, there is an accumulating number of studies reporting increased SOD activities under various biotic and abiotic stress conditions in plants. In this study, eight SOD genes (FSD1-3, CSD1-3 and MSD1-2), under four different abiotic stress conditions, salt, cold, heat and high light exposure, in 10 different Arabidopsis ecotypes produced a total of 320 expression patterns (), of which 142 (∼44%) showed up-regulation, whereas 176 (55%) showed down-regulation. Of the up-regulated genes, 45 ones were induced by salt, 37 by high light, 31 by heat and 29 by cold stress. However, of the down-regulated genes, 51 were induced by cold, 48 by heat, 43 by high light and 34 by salt stress. The up-regulated genes demonstrated a distribution varying in the range of 0.001–1.335 fold change (log2), whereas the down-regulation varied between −0.001 and −2.121 fold. As a criterion of significant expression, we applied the threshold ≥1.0 and ≤ −1.0 log2 fold change. After applying this threshold, a total of 10 conditions, including four up-regulated and six down-regulated genes were identified. Among them, the most up- and down-regulated gene, with 1.335 and −2.121 folds change, was CSD2 under salt (An-1) and cold (Sha) stresses, respectively. Other genes that also demonstrated a significant increase in expression were CSD1 (1.287 fold) under cold stress (Col-0), and FSD1 with 1.195 and 1.171 fold change under heat (Kond) and salt (Kond) stresses, respectively. However, FSD2 showed crucial down-regulation with −1.303 (Cvi) and −1.199 (Kyo-2) fold change under high light, and with −1.155 (Sha) and −1.097 (Kas-1) fold change under cold stress. In addition, MSD1 was also down-regulated with −1.213 folds under cold (Sha) stress. Overall, although all Arabidopsis SOD genes exhibited a varying degree of differential expression, some stresses mostly appeared to induce the chloroplastic FeSOD genes such as FSD1 and 2, and Cu/ZnSOD genes, including cytoplasmic CSD1 and chloroplastic CSD2. Moreover, the regulation of SOD expression did not demonstrate any particular pattern in relation to ecotypes, giving a hint about the different tolerance and response status of each Arabidopsis genotype to given stress factors.
Figure 1. Heatmap showing the expression profiles of eight superoxide dismutase (SOD) genes (CSD1-3, FSD1-3 and MSD1-2) in 10 natural Arabidopsis ecotypes, An-1, Cvi, Col-0, C24, Eri, Kas-1, Kond, Kyo-2, Ler and Sha, under four different stress conditions, salinity (100 mmol/L NaCl), cold (10 °C), heat (38 °C) and high light (800 μmol photons m−2s−1). Green indicates the down-regulated genes; red shows the up-regulated genes under given stresses. Conditions (up) and genes (left) with similar expression profiles were hierarchically clustered using Pearson correlation.
Catalase (EC 1.11.1.6)
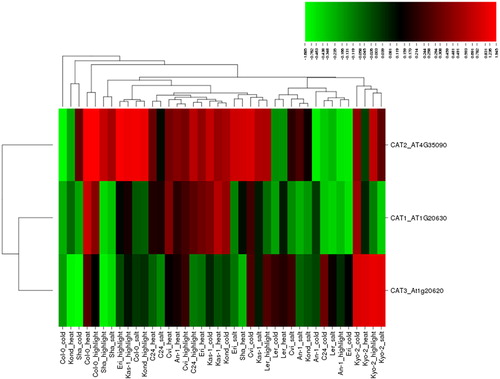
CAT has one of the highest turnover rates of all enzymes: it is estimated that one molecule of CAT is able to convert ∼6 million H2O2 molecules into H2O and O2 per minute [Citation58]. CAT activity has been extensively studied in many higher plants, including maize, barley, Arabidopsis, pumpkin, rice and sunflower [Citation1]. In addition, many studies report the changes in CAT activity under salt, cold, heat and high light stresses, which were evaluated in the present study. For example, CAT activity was reported to increase in chickpea roots in response to salinity [Citation47,Citation49]. In salt-sensitive rice seedlings, CAT activity increased at moderate (7 dS m−1 NaCl) salt treatment, whereas its activity declined with higher salinity (14 dS m−1 NaCl); however in salt-tolerant seedlings, CAT activity was consistently increased by increased salt levels and duration [Citation59]. Changes in CAT activity under salinity have also been reported in many plants, including common bean, Chinese cabbage, pea and maize [Citation60–63]. The cold stress increased the expression of catalase in rice leaf blades [Citation64]. Cold stress was reported to activate the CAT genes in Arabidopsis plants [Citation65]. Transcriptome profiling of cassava plant identified CAT2 among the early cold-responsive genes [Citation66]. High heat stress significantly increased the CAT activity in five wheat genotypes [Citation67]. High CAT activity was observed under high temperature stress in Brassica juncea genotypes but the increase was significantly higher in the tolerant genotype [Citation68]. Under high light, drought stress significantly increased the CAT activity in Picea asperata seedlings [Citation55]. CAT-deficiency was reported to significantly affect the gene expression under high light in Arabidopsis plants [Citation69]. Moreover, many other studies have also reported changes in the activities of CAT enzymes under diverse panels of biotic and abiotic stresses in plants. In the present study, three CAT genes, CAT1-3, under four different abiotic stresses, salt, cold, heat and high light, in 10 Arabidopsis ecotypes produced a total of 120 expression conditions (), of which 76 conditions (∼64%) were up-regulated, while 44 (∼36%) were down-regulated. Of the up-regulations, 22 were induced by heat, 21 by high light, 17 by cold and 16 by salt stress. Besides, 14 of the down-regulations were caused by salt, 13 by cold, 9 by high light and 8 by heat. The up-regulated genes showed a fold change of 0.003–1.945, whereas the down-regulation varied between −0.016 and −1.685. To analyze the significant changes in the expression of the selected CAT genes, we applied a threshold value of ≥1.0 and ≤ −1.0. A total of nine conditions, including six up- and three down-regulated ones, were identified to meet the applied criteria. The most up-regulated gene was CAT2 with 1.945 fold change (Col-0) under high light, whereas the most down-regulated gene was CAT3 with −1.685 fold change (Sha) under cold stress. Interestingly, the expression of CAT2 was also significantly increased in some other cases such as 1.522 fold (Col-0) under heat, 1.274 fold (Col-0) under salt, and 1.385 (Eri), 1.236 (Kas-1) and 1.069 fold (Kond) under high light. However, CAT2 also showed a significant (−1.322 fold) down-regulation under cold stress (Col-0), implicating a stress-specific regulation pattern. In addition, under heat stress, CAT3 exhibited decreased (−1.021 fold) expression (Kond). Although all Arabidopsis catalase genes (CAT1-3) showed some degree of differential expression in response to the studied stress treatments, CAT2 mainly demonstrated a significant up-regulation, whereas CAT3 tended to be down-regulated. The clustering pattern of genes also further supported this, showing that CAT1 and CAT3 genes are grouped together, while CAT2 diverged from them. Moreover, a single ecotype, Kyo-2, exhibited very similar expression patterns of the CAT3 gene under the four stresses, whereas the other genotypes did not show any specific ecotypic distribution, suggesting different stress tolerance of different Arabidopsis ecotypes.
Figure 2. Heatmap showing the expression profiles of three catalase (CAT) genes (CAT1-3) in 10 natural Arabidopsis ecotypes, An-1, Cvi, Col-0, C24, Eri, Kas-1, Kond, Kyo-2, Ler and Sha, under four different stress conditions of salinity (100 mmol/L NaCl), cold (10 °C), heat (38 °C) and high light (800 μmol photons m−2s−1). Green indicates the down-regulated genes; red shows the upregulated genes under given stresses. Conditions (up) and genes (left) with similar expression profiles were hierarchically clustered using Pearson correlation.
Ascorbate peroxidase (EC 1.11.1.11)
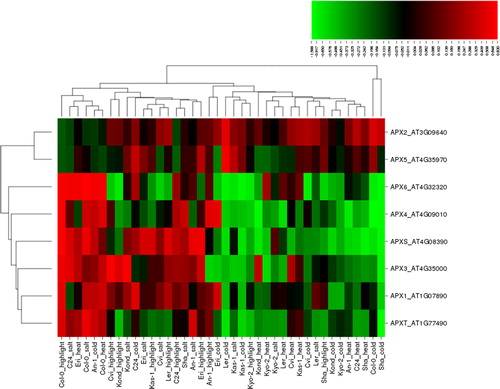
APX has higher affinity for H2O2 (µmol range) than catalase enzymes (mmol range) [Citation1]. In addition, chloroplastic and mitochondrial APXs are reported to less half-inactivation time (<30 sec) than peroxisomal and cytosolic isoforms (≥1 h), therefore making them more sensitive to low level of AsA [Citation70]. Enhanced APX activity in plants has been demonstrated under various abiotic stresses including salt, cold, heat and high light. Studies have reported that salinity increased the transcript levels of APX1, 4, 6 and 7 genes in rice leaf tissues, whereas APX2 expression was not altered but APX8 was slightly decreased [Citation71]. In sorghum, APXs/GPXs genes were mostly up-regulated in leaves, while they were down-regulated in root tissues [Citation72]. Under salinity, salt stress tolerant sweet potato genotypes showed higher APX activity than salt sensitive plants, and APX isoforms demonstrated expression based on tissue type and stress duration [Citation73]. The cytosolic APX2 played a crucial role in rice growth and development by scavenging of ROS under cold, salt and drought stresses [Citation74]. High APX activity improved the recovery of photosynthesis in sugarcane plants under low substrate temperature [Citation75]. APX activity was higher in cold-acclimated barley cultivars than in non-acclimated plants under freezing temperature [Citation76]. The cytosolic APX1 played a crucial role in the acclimation of Arabidopsis plants under combined heat and drought stresses [Citation77]. Heat shock increased the expression of APX2 in leaves of rice seedlings [Citation78]. Increased APX activity in broccoli and Chinese cabbage was reported to play a key role in ROS detoxification under heat stress [Citation79]. Thylakoid APX in Arabidopsis plants protects the photosynthetic apparatus under high light stress by employing the water-water cycle [Citation80]. Arabidopsis APX1 was reported to be involved in H2O2 scavenging under light stress [Citation81]. Excess light in Arabidopsis plants caused a reversible photo-inhibition in photosynthesis, resulting in an increase in the mRNA levels of cytosolic APX1 and APX2 genes [Citation82]. Differential regulation of APX activity has been also demonstrated in many other plants under different biotic/abiotic stress conditions. In the present analysis, eight APX genes, APX1-6, APXT and APXS, under four different stress types, salt, cold, heat and high light, in 10 Arabidopsis ecotypes formed a total of 320 different expression patterns (), of which 149 conditions (∼47%) were up-regulated, while 171 (∼53%) were down-regulated. Among the up-regulated cases, 46 conditions were induced by high light, 43 by salt, 34 by heat and 26 by cold stress; whereas in the 171 down-regulated conditions, 54 were induced by cold, 46 by heat, 37 by salt and 34 by high light. The up-regulation varied in the range of 0.002–0.830 fold change, whereas the down-regulation, between −0.003 and −1.588. As a significant threshold, in this case we applied the cut-off value of ≥0.6 and ≤ −0.6 for differentially expressed genes. A total of 34 conditions, including 13 up- and 21 down-regulated genes were identified by applying this threshold value. APXS was the most up-regulated gene with 0.830 fold change (Col-0) under high light, while APX4 was the most down-regulated gene with −1.588 fold (Col-0) under cold stress. Based on the applied threshold, in some other conditions, the expression of APX3-6 and APXS showed significant up-regulation with 0.603–0.753 fold change. Besides, APX3, APX4, APX6, APXS and APXT demonstrated significant down-regulation ranging between −1.364 and −0.612, particularly, chloroplastic APX4, APX6, APXS and APXT genes under cold (Sha) stress with −1.34, −1.236, −1.14 and −1.364 fold change, respectively. Although all Arabidopsis APX genes were expressed at different levels, chloroplastic APX4, APX6, APXT and APXS and peroxisomal APX3 and APX5 genes appeared to be more responsive under the selected stresses. This is also in agreement with the fact that the chloroplasts and peroxisomes are the main source of ROS production under these stress conditions. Moreover, the gene clustering showed that APX1, APX3, APX4, APX6, APXS and APXT share similar expression profiles. A significant pattern was not observed in relation to the ecotypes, which was indicative of the varying stress tolerance of these Arabidopsis genotypes.
Figure 3. Heatmap showing the expression profiles of eight ascorbate peroxidase (APX) genes (APX1-6, APXT and APXS) in 10 natural Arabidopsis ecotypes, An-1, Cvi, Col-0, C24, Eri, Kas-1, Kond, Kyo-2, Ler and Sha, under four different stress conditions of salinity (100 mmol/L NaCl), cold (10 °C), heat (38 °C) and high light (800 μmol photons m−2s−1). Green indicates the down-regulated genes; red shows the up-regulated genes under given stresses. Conditions (up) and genes (left) with similar expression profiles were hierarchically clustered using Pearson correlation.
Glutathione peroxidase (EC 1.11.1.9)
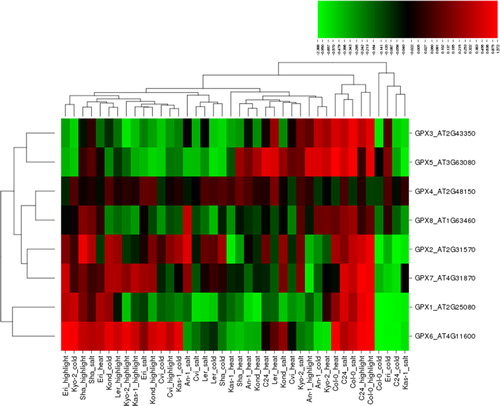
GPX is another important enzymatic antioxidant involved in the reduction of H2O2 as well as organic and lipid hydroperoxides using glutathione (GSH) [Citation1]. Like APX, it helps to protect plants against oxidative stress and its enhanced activity has been reported in many plants under different abiotic stresses, including salt, cold, heat and high light, which the present work aimed to understand in various Arabidopsis ecotypes. Previous studies have reported that salinity stress increased the GPX activity in two rice cultivars [Citation83]. Two proteins related to glutathione-based scavenging of ROS were more abundant in the roots of salt-tolerant barley genotypes, showing increased GPX activity [Citation84]. Salt stress highly up-regulated the expression of GPX in pearl millet, and GPX enhanced the salt tolerance in plants [Citation85]. Cold stress and exogenous H2O2 treatments induced the GPX gene family in rice plants [Citation86]. Transcriptome profiling of cold-tolerant rice plants revealed up-regulated expression of GPX genes under mild cold stress [Citation87]. Chilling-stressed induced RNA-seq analyses showed significant up-regulation of GPX genes in Jatropha curcas [Citation88]. Mung bean seedlings (Vigna radiata L. cv. Binamoog-1) treated with exogenous GSH under high temperature increased the GPX activity and showed enhanced high temperature stress tolerance [Citation89]. Heat treatment was reported to increase the GPX activity in T. aestivum seedlings [Citation90]. Heat stress-induced transcriptome analysis revealed the increased GPX activity in switchgrass plants [Citation91]. Hairy root culture of wild carrot under light irritation demonstrated the reduced GPX activity [Citation92]. Expression analysis revealed that the cucumber GPX genes showed different expression patterns under abscisic acid treatments and abiotic stress conditions [Citation93]. In addition, most of the TaGPX genes indicated higher expression levels in various leaf developmental stages under abiotic stress conditions in wheat [Citation94]. Abiotic stress-induced GPX activity has been also reported in many other plants, showing the importance of GPX members in ROS-scavenging. In this work, eight GPX genes, GPX1-8, in 10 Arabidopsis ecotypes under the four different stresses that we focussed on, resulted in a total of 320 different expression profiles (): 142 (∼45%) up-regulated and 177 (∼55%) down-regulated conditions. Of the up-regulated conditions, 43 were induced by high light, 42 by salt, 29 by heat and 28 by cold stress, whereas of the down-regulated conditions, 52 were caused by cold, 51 by heat, 38 by salt and 36 by high light. The up-regulated genes showed a variation of 0.005–1.572 fold change, and the down-regulated genes varied between −2.388 and −0.001. Genes with significant differential expression were determined based on a threshold value of ≥1.0 and ≤ −1.0. This cut-off value identified a total of 10 conditions, including four up- and six down-regulated ones. Arabidopsis GPX6 was identified as the gene with the most pronounced differential expression: it was up- and down-regulated by 1.572 and −2.388 fold (Col-0) under high light and cold stresses, respectively. Thus, the expression of mitochondrial GPX6 could be considered the most responsive gene under this type of stress. It also implies that the most likely source of ROS are mitochondria. Other significantly up-regulated genes were GPX5 with 1.289-fold (Col-0) under heat, and GPX6 and 7 with 1.15 fold (Eri) and 1.112 fold (C24) under high light. In contrast, some genes showed down-regulation under cold stress: GPX1 with −2.152 (Col-0) and −1.248 fold (Eri), GPX6 with −1.216 (Eri) and −1.062 fold (C24), and GPX2 with −1.049 fold (C24), indicating that cold stress may appear to significantly down-regulate the expression of some antioxidant genes. Moreover, chloroplastic GPX1 and mitochondrial GPX6 demonstrated similar expression patterns; other genes that shared similar expression patters were GPX2, GPX3, GPX4, GPX5, GPX7 and GPX8. Furthermore, the lack of any obvious pattern in the clustering of the ecotypes under these stresses might be attributed to different stress tolerance of these Arabidopsis genotypes.
Figure 4. Heatmap showing the expression profiles of eight glutathione peroxidase (GPX) genes (GPX1-8) in 10 natural Arabidopsis ecotypes, An-1, Cvi, Col-0, C24, Eri, Kas-1, Kond, Kyo-2, Ler and Sha, under four different stress conditions of salinity (100 mmol/L NaCl), cold (10 °C), heat (38 °C) and high light (800 μmol photons m−2s−1). Green indicates the down-regulated genes; red shows the up-regulated genes under given stresses. Conditions (up) and genes (left) with similar expression profiles were hierarchically clustered using Pearson correlation.
Monodehydroascorbate reductase (EC 1.6.5.4)
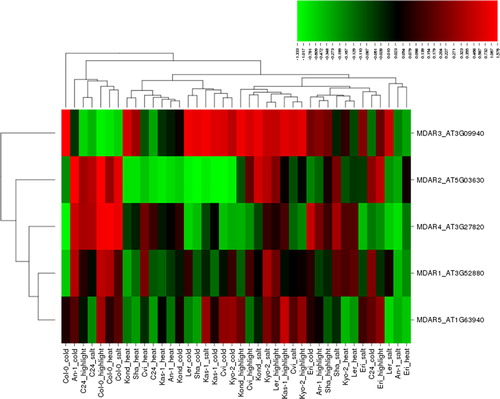
MDHAR protects against ROS via involvement in the ascorbate-glutathione (ASH-GSH) cycle, maintaining ascorbate (AsA) in a reduced state in cellular compartments [Citation1,Citation95]. A substantial number of studies have reported the involvement of MDHAR in plant protection against ROS under various abiotic stresses, including salt, cold, heat and high light. For example, Truffault et al. [Citation96] reported that MDHAR activity could be a potential candidate to enhance the yield in crops under drought or light stress. NaCl-salt stress (300 mmol/L) significantly up-regulated the cytosolic MDHAR in Aeluropus littoralis shoots and roots but MDHAR accumulation declined at concentrations higher than 300 mmol/L [Citation97]. Reduced MDHAR activity decreased the cold storage tolerance in tomato plants and affected the antioxidant levels in fruits [Citation98]. MDHAR activity was significantly inhibited at either high or low light following cold stress [Citation99]. Prolonged exposure to heat stress increased the transcript levels of MDHAR in rice leaves [Citation100]. MDHAR activity was higher in short-term (3 days) heat stress but its activity gradually decreased in long-term treatment [Citation101]. Similar results were also reported in a different study, in which heat stress significantly increased the MDHAR activity in a grass species, Poa pratensis, but its activity declined under long-term exposure [Citation102]. In DHAR3 Arabidopsis mutants, high light stress activated the MDHAR gene to compensate the DHAR3 activity [Citation103]. MDHAR is reportedly involved in the regulation of ascorbate in tomato plants in a light-dependent manner [Citation104]. MDHAR activity was associated with sugar levels in tomato under high light, and its reduced activity inhibited the plant growth and yield [Citation96]. The involvement of MDHAR in ROS-regulation, thereby in plant stress tolerance, has been also demonstrated in many other studies performed under various stress conditions. In this study, five MDHAR genes, MDAR1-5, under four different stresses such as salt, cold, heat and high light in 10 Arabidopsis ecotypes produced a total of 200 differentially expressed patterns (), of which 110 (55%) showed up-regulation, while 89 (∼45%) down-regulation. Among the up-regulated conditions, there were 36 ones induced by high light, 29 by cold, 27 by salt and 18 by heat stress. Among the down-regulated conditions, 31 were induced by heat, 23 by salt, 21 by cold and 14 by high light. The extent of up-regulation varied within 0.010–1.576 fold change, while that of down-regulation, between −1.333 and −0.006. To identify the genes whose expression levels were significantly altered, we adopted a threshold range of ≥1.0 and ≤ −1.0. This threshold identified 15 conditions, including eight up- and seven down-regulated ones. Arabidopsis MDAR3 was the most up-regulated gene with 1.576 fold change (Col-0), while MDAR2 was the most down-regulated one with −1.333 fold change (Sha) under cold stress. There was also significant up-regulation of MDAR2 with 1.19 fold (An-1) and 1.17 fold (Col-0) under cold and high light stresses, respectively; MDAR3 with 1.14 (Kas-1), 1.067 (Ler) and 1.041 fold (Kond) under salt, cold and high light, respectively; and MDAR4 with 1.146 and 1.144 fold (Col-0) under heat and high light, respectively. However, significant down-regulation was observed for MDAR2 by −1.274 fold (Cvi), −1.224 (Kas-1), −1.098 (Ler) and −1.017 fold (Kyo-2) under cold stress; MDAR3 with −1.023 fold (Col-0) under high light; and MDAR4 with −1.205 (Col-0) under cold stress. As such, the cytosolic MDAR2 and MDAR3 and the peroxisomal MDAR4 were the most responsive genes under these stresses, particularly under cold stress. Moreover, MDAR2-4 demonstrated different expression patterns in clustering, while MDAR1 and MDAR5 had similar expression. Furthermore, the clustering pattern of ecotypes under these stresses also suggested different stress tolerance of the Arabidopsis genotypes included in this study.
Figure 5. Heatmap showing the expression profiles of five monodehydroascorbate reductase (MDHAR) genes (MDAR1-5) in 10 natural Arabidopsis ecotypes, An-1, Cvi, Col-0, C24, Eri, Kas-1, Kond, Kyo-2, Ler and Sha, under four different stress conditions of salinity (100 mmol/L NaCl), cold (10 °C), heat (38 °C) and high light (800 μmol photons m−2s−1). Green indicates the down-regulated genes; red shows the up-regulated genes under given stresses. Conditions (up) and genes (left) with similar expression profiles were hierarchically clustered using Pearson correlation.
Dehydroascorbate reductase (EC 1.8.5.1)
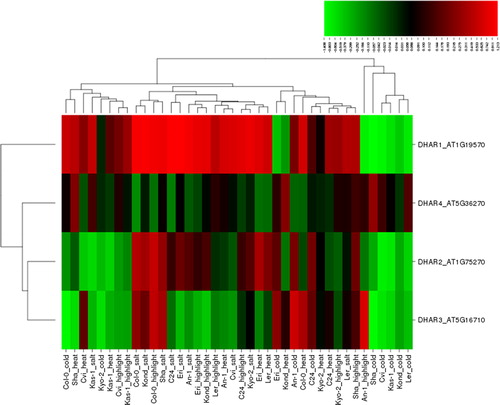
DHAR maintains the redox pools of AsA through recycling of oxidized AsA to reduced AsA. Thus, it is of crucial significance in the regulation of ROS-scavenging in plants, especially under stress conditions [Citation1,Citation39]. The important role of DHAR in ROS detoxification has been widely reported under various abiotic stresses. For example, salt stress treatment decreased the DHAR activity in cucumber [Citation105]. In salt-sensitive rice seedlings, the DHAR activity increased at moderate salinity (7 dS m−1 NaCl) but declined with higher salinity (14 dS m−1) [Citation55]. High or low light following cold stress significantly inhibited the DHAR activity in cucumber (Cucumis sativus) [Citation99]. RNA-Seq analysis revealed up-regulation of the DHAR gene in cold-induced Jatropha curcas [Citation88]. DHAR exhibited higher activity in short-term (3 days) heat stress but declined activity in further treatments [Citation101]. Upon heat stress, the DHAR gene demonstrated an increased transcript level in rice leaves [Citation100]. The DHAR activity significantly increased in heat-tolerant grass species, Poa pratensis under heat stress but its activity declined under long-term exposure [Citation102]. High light stress induced the MDHAR gene in DHAR3 Arabidopsis mutants, compensating DHAR3 activity [Citation103]. Proteomic analysis showed increased DHAR activity in Arabidopsis chloroplasts under high light stress [Citation106]. Under high light, the transcript level of DHAR markedly increased in salt-stressed tomato fruits [Citation107]. Many other studies also report on the significant role of DHAR in plant stress tolerance under various stresses. In the present study, four DHAR genes, DAR1-4, under four different abiotic stress conditions, salt, cold, heat and high light, in 10 Arabidopsis ecotypes gave a total of 160 different expression patterns (), of which 95 (59%) displayed up-regulation, while 65 (∼41%) down-regulation. Among the up-regulated conditions, 30 were induced by salt, 25 by high light, 22 by heat and 18 by cold stress. Among the down-regulated conditions, 22 were induced by cold, 18 by heat, 15 by high light and 10 by salt stress. The up-regulated genes showed a distribution with 0.007–1.213 fold change, whereas the down-regulated ones, between −1.408 and −0.004 fold. Genes with significantly differential expression were determined based on a cut-off value of ≥1.0 and ≤ −1.0. According to this threshold, six conditions, including four up- and two down-regulated ones showed the significant expression. Arabidopsis DHAR1 was identified to be the most up- and down-regulated gene with 1.213 (Eri) and −1.408 fold change (Sha) under salt and cold stress, respectively. DHAR1 was significantly up-regulated with 1.199 (C24), 1.155 (Kond) and 1.148 fold change (Col-0) under salt stress, whereas DHAR3 with -1.364 fold change (Sha) under cold stress was considerably down-regulated. Thus, the mitochondrial DHAR1 and the chloroplastic DHAR3 appeared to be the most responsive genes under the selected abiotic stresses, especially in response to cold and salt stress. Moreover, the hierarchical clustering of DHAR2-4 genes under these stresses showed a more similar expression pattern than DHAR1. Furthermore, although some ecotypes shared similar expression patterns for particular stress types, the overall clustering pattern under the selected stresses mainly indicated different stress tolerance of the Arabidopsis genotypes.
Figure 6. Heatmap showing the expression profiles of four dehydroascorbate reductase (DHAR) genes (DHAR1-4) in 10 natural Arabidopsis ecotypes, An-1, Cvi, Col-0, C24, Eri, Kas-1, Kond, Kyo-2, Ler and Sha, under four different stress conditions of salinity (100 mmol/L NaCl), cold (10 °C), heat (38 °C) and high light (800 μmol photons m−2s−1). Green indicates the down-regulated genes; red shows the up-regulated genes under given stresses. Conditions (up) and genes (left) with similar expression profiles were hierarchically clustered using Pearson correlation.
Overall, the expression profiles of the major Arabidopsis antioxidant genes varied based on stress and enzyme type and plant ecotype. However, among them, the ones that showed the most pronounced up- and down-regulation respectively included CSD2 (An-1; salt; 1.335 fold) and CSD2 (Sha; cold; -2.121 fold) in SODs; CAT2 (Col-0; high light; 1.945 fold) and CAT3 (Sha; cold; −1.685 fold) in CATs; APXS (Col-0; high light; 0.830 folds) and APX4 (Col-0; cold; −1.588 fold) in APXs; GPX6 (Col-0; high light; 1.572 fold) and GPX6 (Col-0; cold; −2.388 fold) in GPXs; MDAR3 (Col-0; cold; 1.576 fold) and MDAR2 (Sha; cold; −1.333 fold) in MDHARs; and DHAR1 (Eri; salt; 1.213 fold) and DHAR1 (Sha; cold; −1.408 fold) in DHARs. In terms of stress type, mainly high light as well as salt and cold treatment significantly up-regulated the gene expression, whereas cold stress caused mainly down-regulation. The Arabidopsis ecotypes showed different levels of stress tolerance. The pronounced expression of the genes from the SOD, CAT, APX, GPX, MDHAR and DHAR families under given conditions highlighted their crucial roles in ROS-scavenging under abiotic stress.
Conclusions
Taking advantage of microarray data, the expression profiles of six Arabidopsis antioxidant enzyme genes encoding SOD, CAT, APX, GPX, MDHAR and DHAR, under four different abiotic stress conditions, cold, heat, high light and salt, were evaluated in 10 natural Arabidopsis ecotypes originating from different geographical locations. A total of 36 gene isoforms, including CSD1-3, FSD1-3, MSD1-2, CAT1-3, APX1-6, APXT, APXS, GPX1-8, MDAR1-5 and DHAR1-4, were investigated. Under these stresses, CAT genes had the most up-regulated expression with about 64% of the conditions, whereas SOD and GPX demonstrated the most down-regulated expression with about 55% of the conditions. The expression levels of the up-regulated antioxidant genes ranged from 0.001 to 1.945 fold change, whereas the expression of the down-regulated genes varied between −0.001 and −2.388 fold. Among the enzyme isoforms, CAT2 showed the highest increase by 1.945 fold under high light in Col-0 ecotype, followed by MDAR3 with 1.576 fold (Col-0) under cold stress, GPX6 with 1.572 fold (Col-0) under high light, CSD2 and DHAR1 with 1.335 (An-1) and 1.213 (Eri) fold under salt stress, respectively, and APXS with 0.830 fold (Col-0) under high light exposure. As such, the types of abiotic stresses that considerably up-regulated the gene expression were high light as well as salt and cold stress, implicating that Arabidopsis ecotypes could be more vulnerable to photo-oxidative damage. However, GPX6 was the most down-regulated antioxidant gene, with −2.388 fold change under cold stress in Col-0 ecotype, followed by CSD2 with −2.121 (Sha), CAT3 with −1.685 (Sha), APX4 with −1.588 (Col-0), DHAR1 with −1.408 (Sha) and MDAR2 with −1.333 fold change (Sha) under cold stress. Thus, cold stress seemed to exert down-regulation of some of the analyzed genes. In addition, differential expression of Sha and Col-0 genotypes suggested that these varieties could be more sensitive to given stresses. However, overall the expression pattern of the ecotypes suggested different stress tolerance of these Arabidopsis genotypes. Moreover, Arabidopsis FSD1, FSD2, CSD1 and CSD2 genes from the SOD family; CAT2 and CAT3 from CAT; APX3-6, APXT and APXS from APX; GPX1, GPX2, GPX5, GPX6 and GPX7 from GPX; MDAR2-4 from MDHAR; and DHAR1 and DHAR3 from the DHAR family appeared to be more differentially expressed under given stress conditions, emphasizing their crucial roles in ROS-scavenging. Thus, the results from this study provide insight into better understanding the enzymatic antioxidant mechanisms in plants, with an ultimate goal of engineering abiotic stress tolerant crops.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Gill SS, Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem. 2010;48:909–930.
- Petrov VD, Van Breusegem F. Hydrogen peroxide-a central hub for information flow in plant cells. AoB Plants. 2012;2012:pls014. DOI: 10.1093/aobpla/pls014
- Fam SS, Morrow JD. The isoprostanes: unique products of arachidonic acid oxidation-a review. Curr Med Chem. 2003;10:1723–1740.
- Montillet JL, Chamnongpol S, Rustérucci C, et al. Fatty acid hydroperoxides and H2O2 in the execution of hypersensitive cell death in tobacco leaves. Plant Physiol. 2005;138:1516–1526.
- Garg N, Manchanda G. ROS generation in plants: boon or bane? Plant Biosys. 2009;143:81–96.
- Møller IM, Jensen PE, Hansson A. Oxidative modifications to cellular components in plants. Annu Rev Plant Biol. 2007;58:459–481.
- Tuteja N, Singh MB, Misra MK, et al. Molecular mechanisms of DNA damage and repair: progress in plants. Crit Rev Biochem Mol Biol. 2001;36:337–397.
- Britt AB. Molecular genetics of DNA repair in higher plants. Trends Plant Sci. 1999;4:20–25.
- Cooke MS, Evans MD, Dizdaroglu M, et al. Oxidative DNA damage: mechanisms, mutation, and disease. Faseb J. 2003;17:1195–1214.
- Wagner D, Przybyla D, Op den Camp R, et al. The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science. 2004;306:1183–1185.
- Halliwell B. Reactive species and antioxidants. Redox biology is a fundamental theme of aerobic life. Plant Physiol. 2006;141:312–322.
- Tewari RK, Kumar P, Sharma PN. Antioxidant responses to enhanced generation of superoxide anion radical and hydrogen peroxide in the copper-stressed mulberry plants. Planta. 2006;223:1145–1153.
- Luis A, Sandalio LM, Corpas FJ, et al. Reactive oxygen species and reactive nitrogen species in peroxisomes. Production, scavenging, and role in cell signaling. Plant Physiol. 2006;141:330–335.
- Navrot N, Rouhier N, Gelhaye E, et al. Reactive oxygen species generation and antioxidant systems in plant mitochondria. Physiol Plant. 2007;129:185–195.
- Bolwell GP, Wojtaszek P. Mechanisms for the generation of reactive oxygen species in plant defence–a broad perspective. Physiol Mol Plant Pathol. 1997;51:347–366.
- Foyer CH, Noctor G. Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell. 2005;17:1866–1875.
- Sharma P, Jha AB, Dubey RS, et al. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J Bot. 2012;2012:217037. DOI:10.1155/2012/217037
- Quan LJ, Zhang B, Shi WW, et al. Hydrogen peroxide in plants: a versatile molecule of the reactive oxygen species network. J Integr Plant Biol. 2008;50:2–18.
- Foreman J, Demidchik V, Bothwell JH, et al. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature. 2003;422:442–446.
- Mittler R, Vanderauwera S, Gollery M, et al. Reactive oxygen gene network of plants. Trends Plant Sci. 2004;9:490–498.
- Noctor G, Foyer CH. A re-evaluation of the ATP: NADPH budget during C3 photosynthesis: a contribution from nitrate assimilation and its associated respiratory activity? J Expr Bot. 1998;49:1895–1908.
- Bright J, Desikan R, Hancock JT, et al. ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. Plant J. 2006;45:113–122.
- Ozyigit II, Filiz E, Vatansever R, et al. Identification and comparative analysis of H2O2-scavenging enzymes (ascorbate peroxidase and glutathione peroxidase) in selected plants employing bioinformatics approaches. Front Plant Sci. 2016;7:301. DOI: 10.3389/fpls.2016.00301
- Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002;7:405–410.
- Kliebenstein DJ, Dietrich RA, Martin AC, et al. LSD1 regulates salicylic acid induction of copper zinc superoxide dismutase in Arabidopsis thaliana. Mol Plant Microbe Interact. 1999;12:1022–1026.
- Azevedo RA, Alas RM, Smith RJ, et al. Response of antioxidant enzymes to transfer from elevated carbon dioxide to air and ozone fumigation, in the leaves and roots of wild‐type and a catalase‐deficient mutant of barley. Physiol Plant. 1998;104:280–292.
- Frugoli JA, Zhong HH, Nuccio ML, et al. Catalase is encoded by a multigene family in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 1996;112:327–336.
- Guan L, Scandalios JG. Molecular evolution of maize catalases and their relationship to other eukaryotic and prokaryotic catalases. J Mol Evol. 1996;42:570–579.
- Esaka M, Yamada N, Kitabayashi M, et al. cDNA cloning and differential gene expression of three catalases in pumpkin. Plant Mol Biol. 1997;33:141–155.
- Iwamoto M, Higo H, Higo K. Differential diurnal expression of rice catalase genes: the 5′-flanking region of CatA is not sufficient for circadian control. Plant Sci. 2000;151:39–46.
- Azpilicueta CE, Benavides MP, Tomaro ML, et al. Mechanism of CATA3 induction by cadmium in sunflower leaves. Plant Physiol Biochem. 2007;45:589–595.
- Teixeira FK, Menezes-Benavente L, Margis R, et al. Analysis of the molecular evolutionary history of the ascorbate peroxidase gene family: inferences from the rice genome. J Mol Evol. 2004;59:761–770.
- Panchuk II, Zentgraf U, Volkov RA. Expression of the Apx gene family during leaf senescence of Arabidopsis thaliana. Planta. 2005;222:926–932.
- Najami N, Janda T, Barriah W, et al. Ascorbate peroxidase gene family in tomato: its identification and characterization. Mol Genet Genomics.. 2008;279:171–182.
- Milla MAR, Maurer A, Huete AR, et al. Glutathione peroxidase genes in Arabidopsis are ubiquitous and regulated by abiotic stresses through diverse signaling pathways. Plant J. 2003;36:602–615.
- Koua D, Cerutti L, Falquet L, et al. PeroxiBase: a database with new tools for peroxidase family classification. Nucleic Acids Res. 2009;37:D261–D266.
- Yoon HS, Lee H, Lee IA, et al. Molecular cloning of the monodehydroascorbate reductase gene from Brassica campestris and analysis of its mRNA level in response to oxidative stress. Biochim Biophys Acta. 2004;1658:181–186.
- Bodra N, Young D, Rosado LA, et al. Arabidopsis thaliana dehydroascorbate reductase 2: conformational flexibility during catalysis. Sci Rep. 2017;7:42494. DOI: 10.1038/srep42494
- Noshi M, Yamada H, Hatanaka R, et al. Arabidopsis dehydroascorbate reductase 1 and 2 modulate redox states of ascorbate-glutathione cycle in the cytosol in response to photooxidative stress. Biosci Biotechnol Biochem. 2017;81:523–533.
- Trivedi DK, Gill SS, Yadav S, et al. Genome-wide analysis of glutathione reductase (GR) genes from rice and Arabidopsis. Plant Signal Behav. 2013;8:e23021. DOI: 10.4161/psb.23021
- Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55:373–399.
- Reade JPH. Plant stress biology: from genomics to system biology, edited by H. Hirt. XVI +257 pp. Weinheim: Wiley-VCH Verlag GmbH & Co. KGaA (2009). ISBN: 978-3-527-32290-9. J Agr Sci. 2010;149:125.
- Rasmussen S, Barah P, Suarez-Rodriguez MC, et al. Transcriptome responses to combinations of stresses in Arabidopsis. Plant Physiol. 2013;161:1783–1794.
- Barah P, B N MN, Jayavelu ND, et al. Transcriptional regulatory networks in Arabidopsis thaliana during single and combined stresses. Nucleic Acids Res. 2016;44:3147–3164.
- Scherf U, Ross DT, Waltham M, et al. A gene expression database for the molecular pharmacology of cancer. Nat Genet. 2000;24:236–244.
- Harinasut P, Poonsopa D, Roengmongkol K, et al. Salinity effects on antioxidant enzymes in mulberry cultivar. Sci Asia. 2003;29:109–113.
- Kukreja S, Nandwal AS, Kumar N, et al. Plant water status, H2O2 scavenging enzymes, ethylene evolution and membrane integrity of Cicer arietinum roots as affected by salinity. Biol Plant. 2005;49:305–308.
- Gapińska M, Skłodowska M, Gabara B. Effect of short-and long-term salinity on the activities of antioxidative enzymes and lipid peroxidation in tomato roots. Acta Physiol Plant. 2007;30:11–18.
- Eyidogan F, Oz MT. Effect of salinity on antioxidant responses of chickpea seedlings. Acta Physiol Plant. 2007;29:485–493.
- Pan Y, Wu LJ, Yu ZL. Effect of salt and drought stress on antioxidant enzymes activities and SOD isoenzymes of liquorice (Glycyrrhiza uralensis Fisch). Plant Growth Regul. 2006;49:157–165.
- Zhu Z, Jiang JY, Jiang CJ, et al. Effects of low temperature stress on SOD activity, soluble protein content and soluble sugar content in Camellia sinensis leaves. J Anhui Agr Uni. 2011;1:24–26.
- Nayyar H, Chander S. Protective effects of polyamines against oxidative stress induced by water and cold stress in chick pea. J Agron Crop Sci. 2004;190:355–365.
- Seppänen MM, Fagerstedt K. The role of superoxide dismutase activity in response to cold acclimation in potato. Physiol Plant. 2000;108:279–285.
- Cakmak I, Marschner H. Magnesium deficiency and high light intensity enhance activities of superoxide dismutase, ascorbate peroxidase, and glutathione reductase in bean leaves. Plant Physiol. 1992;98:1222–1227.
- Yang Y, Han C, Liu Q, et al. Effect of drought and low light on growth and enzymatic antioxidant system of Picea asperata seedlings. Acta Physiol Plant. 2008;30:433–440.
- Wang W, Zhang X, Deng F, et al. Genome-wide characterization and expression analyses of superoxide dismutase (SOD) genes in Gossypium hirsutum. BMC Genomics. 2017;18:376. Doi: 10.1186/s12864-017-3768-5
- Chaitanya KV, Sundar D, Masilamani S, et al. Variation in heat stress-induced antioxidant enzyme activities among three mulberry cultivars. Plant Growth Regul. 2002;36:175–180.
- Tanveer M, Shabala S, et al. Targeting redox regulatory mechanisms for salinity stress tolerance in crops. In: Kumar V, Wani S, Suprasanna P, editors. Salinity responses and tolerance in plants, Vol 1. Cham: Springer; 2018. p. 213–234.
- Mishra P, Bhoomika K, Dubey RS. Differential responses of antioxidative defense system to prolonged salinity stress in salt-tolerant and salt-sensitive Indica rice (Oryza sativa L.) seedlings. Protoplasma. 2013;250:3–19.
- Hernández JA, Almansa MS. Short-term effects of salt stress on antioxidant systems and leaf water relations of pea leaves. Physiol Plant. 2002;115:251–257.
- Jebara S, Jebara M, Limam F, et al. Changes in ascorbate peroxidase, catalase, guaiacol peroxidase and superoxide dismutase activities in common bean (Phaseolus vulgaris) nodules under salt stress. J Plant Physiol. 2005;162:929–936.
- Tseng MJ, Liu CW, Yiu JC. Enhanced tolerance to sulfur dioxide and salt stress of transgenic Chinese cabbage plants expressing both superoxide dismutase and catalase in chloroplasts. Plant Physiol Biochem. 2007;45:822–833.
- Gondim FA, Gomes-Filho E, Costa JH, et al. Catalase plays a key role in salt stress acclimation induced by hydrogen peroxide pretreatment in maize. Plant Physiol Biochem. 2012;56:62–71.
- Hashimoto M, Komatsu S. Proteomic analysis of rice seedlings during cold stress. Proteomics. 2007;7:1293–1302.
- Du YY, Wang PC, Chen J, et al. Comprehensive functional analysis of the catalase gene family in Arabidopsis thaliana. J Integr Plant Biol. 2008;50:1318–1326.
- An D, Yang J, Zhang P. Transcriptome profiling of low temperature-treated cassava apical shoots showed dynamic responses of tropical plant to cold stress. BMC Genomics. 2012;13:64. DOI: 10.1186/1471-2164-13-64
- Almeselmani M, Deshmukh PS, Sairam RK, et al. Protective role of antioxidant enzymes under high temperature stress. Plant Sci. 2006;171:382–388.
- Rani B, Dhawan K, Jain V, et al. High temperature induced changes in antioxidative enzymes in Brassica juncea (L) Czern&Coss. 2013. [cited 2018 Apr 05]; [12:1–4]. Available from: http://www.australianoilseeds.com.
- Vandenabeele S, Vanderauwera S, Vuylsteke M, et al. Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant J. 2004;39:45–58.
- Caverzan A, Passaia G, Rosa SB, et al. Plant responses to stresses: role of ascorbate peroxidase in the antioxidant protection. Genet Mol Biol. 2012;35:1011–1019.
- Yamane K, Mitsuya S, Taniguchi M, et al. Transcription profiles of genes encoding catalase and ascorbate peroxidase in the rice leaf tissues under salinity. Plant Prod Sci. 2010;13:164–168.
- Akbudak MA, Filiz E, Vatansever R, et al. Genome-wide identification and expression profiling of ascorbate peroxidase (APX) and glutathione peroxidase (GPX) genes under drought stress in sorghum (Sorghum bicolor L.). J Plant Growth Regul. 2018;37:925–936.
- Lin KH, Pu SF. Tissue-and genotype-specific ascorbate peroxidase expression in sweet potato in response to salt stress. Biol Plant. 2010;54:664–670.
- Zhang Z, Zhang Q, Wu J, et al. Gene knockout study reveals that cytosolic ascorbate peroxidase 2 (OsAPX2) plays a critical role in growth and reproduction in rice under drought, salt and cold stresses. PLoS One. 2013;8:e57472. DOI: 10.1371/journal.pone.0057472
- Sales CR, Ribeiro RV, Silveira JA, et al. Superoxide dismutase and ascorbate peroxidase improve the recovery of photosynthesis in sugarcane plants subjected to water deficit and low substrate temperature. Plant Physiol Biochem. 2013;73:326–336.
- Dai F, Huang Y, Zhou M, et al. The influence of cold acclimation on antioxidative enzymes and antioxidants in sensitive and tolerant barley cultivars. Biol Plant. 2009;53:257–262.
- Koussevitzky S, Suzuki N, Huntington S, et al. Ascorbate peroxidase 1 plays a key role in the response of Arabidopsis thaliana to stress combination. J Biol Chem. 2008; 283:34197–34203.
- Chou TS, Chao YY, Kao CH. Involvement of hydrogen peroxide in heat shock-and cadmium-induced expression of ascorbate peroxidase and glutathione reductase in leaves of rice seedlings. J Plant Physiol. 2012;169:478–486.
- Lin KH, Huang HC, Lin CY. Cloning, expression and physiological analysis of broccoli catalase gene and Chinese cabbage ascorbate peroxidase gene under heat stress. Plant Cell Rep. 2010;29:575–593.
- Awad J, Stotz HU, Fekete A, et al. 2-Cysteine peroxiredoxins and thylakoid ascorbate peroxidase create a water-water cycle that is essential to protect the photosynthetic apparatus under high light stress conditions. Plant Physiol. 2015;167:1592–1603.
- Suzuki N, Devireddy AR, Inupakutika MA, et al. Ultra‐fast alterations in mRNA levels uncover multiple players in light stress acclimation in plants. Plant J. 2015;84:760–772.
- Karpinski S, Escobar C, Karpinska B, et al. Photosynthetic electrontransport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell. 1997;9:627–640.
- Khan MH, Panda SK. Alterations in root lipid peroxidation and antioxidative responses in two rice cultivars under NaCl-salinity stress. Acta Physiol Plant. 2007;30:81–89.
- Witzel K, Weidner A, Surabhi GK, et al. Salt stress-induced alterations in the root proteome of barley genotypes with contrasting response towards salinity. J Expr Bot. 2009;60:3545–3557.
- Islam T, Manna M, Reddy MK. Glutathione peroxidase of Pennisetum glaucum (PgGPx) is a functional Cd2+ dependent peroxiredoxin that enhances tolerance against salinity and drought stress. PLoS One. 2015;10:e0143344. DOI: 10.1371/journal.pone.0143344
- Passaia G, Fonini LS, Caverzan A, et al. The mitochondrial glutathione peroxidase GPX3 is essential for H2O2 homeostasis and root and shoot development in rice. Plant Sci. 2013;208:93–101.
- Zhao J, Zhang S, Yang T, et al. Global transcriptional profiling of a cold‐tolerant rice variety under moderate cold stress reveals different cold stress response mechanisms. Physiol Plantarum. 2015;154:381–394.
- Wang H, Zou Z, Wang S, et al. Deep sequencing-based transcriptome analysis of the oil-bearing plant Physic Nut (Jatropha curcas L.) under cold stress. Plant Omics. 2014;7:178–187.
- Nahar K, Hasanuzzaman M, Alam MM, et al. Exogenous glutathione confers high temperature stress tolerance in mung bean (Vigna radiata L.) by modulating antioxidant defense and methylglyoxal detoxification system. Environ Expr Bot. 2015;112:44–54.
- Hasanuzzaman M, Nahar K, Alam MM, et al. Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int J Mol Sci. 2013;14:9643–9684.
- Li YF, Wang Y, Tang Y, et al. Transcriptome analysis of heat stress response in switchgrass (Panicum virgatum L.). BMC Plant Biol. 2013;13:153. DOI: 10.1186/1471-2229-13-153
- Mukherjee C, Sircar D, Chatterjee M, et al. Combating photooxidative stress in green hairy roots of Daucus carota cultivated under light irradiation. J Plant Physiol. 2014;171:179–187.
- Zhou Y, Hu L, Ye S, et al. Genome-wide identification of glutathione peroxidase (GPX) gene family and their response to abiotic stress in cucumber. 3 Biotech. 2018;8:159. DOI: 10.1007/s13205-018-1185-3
- Tyagi S, Sembi JK, Upadhyay SK. Gene architecture and expression analyses provide insights into the role of glutathione peroxidases (GPXs) in bread wheat (Triticum aestivum L.). J Plant Physiol. 2018;223:19–31.
- Feng H, Liu W, Zhang Q, et al. TaMDHAR4, a monodehydroascorbate reductase gene participates in the interactions between wheat and Puccinia striiformis f. sp. tritici. Plant Physiol Biochem. 2014;76:7–16.
- Truffault V, Gest N, Garchery C, et al. Reduction of MDHAR activity in cherry tomato suppresses growth and yield and MDHAR activity is correlated with sugar levels under high light. Plant Cell Environ. 2016;39:1279–1292.
- Mohseni A, Nematzadeh GA, Dehestani A, et al. Isolation, molecular cloning and expression analysis of Aeluropus littoralis Monodehydroascorbate reductase (MDHAR) gene under salt stress. J Plant Mol Breed. 2015;3:72–80.
- El Airaj H, Gest N, Truffault V, et al. Decreased monodehydroascorbate reductase activity reduces tolerance to cold storage in tomato and affects fruit antioxidant levels. Postharvest BiolTec. 2013;86:502–510.
- Jiang YP, Huang LF, Cheng F, et al. Brassinosteroids accelerate recovery of photosynthetic apparatus from cold stress by balancing the electron partitioning, carboxylation and redox homeostasis in cucumber. Physiol Plantarum. 2013;148:133–145.
- Lee DG, Ahsan N, Kim YG, et al. Expression of heat shock protein and antioxidant genes in rice leaf under heat stress. J Kor Soc Grassland Forage Sci. 2013;33:159–166.
- Sgobba A, Paradiso A, Dipierro S, et al. Changes in antioxidants are critical in determining cell responses to short- and long-term heat stress. Physiol Plant. 2015;153:68–78.
- Du H, Zhou P, Huang B. Antioxidant enzymatic activities and gene expression associated with heat tolerance in a cool-season perennial grass species. Environ Exp Bot. 2013;87:159–166.
- Noshi M, Hatanaka R, Tanabe N, et al. Redox regulation of ascorbate and glutathione by a chloroplastic dehydroascorbate reductase is required for high-light stress tolerance in Arabidopsis. Biosci Biotech Biochem. 2016;80:870–877.
- Gest N, Garchery C, Gautier H, et al. Light‐dependent regulation of ascorbate in tomato by a monodehydroascorbate reductase localized in peroxisomes and the cytosol. Plant Biotechnol J. 2013;11:344–354.
- Shu S, Yuan LY, Guo SR, et al. Effects of exogenous spermine on chlorophyll fluorescence, antioxidant system and ultrastructure of chloroplasts in Cucumis sativus L. under salt stress. Plant Physiol Biochem. 2013;63:209–216.
- Phee BK, Cho JH, Park S, et al. Proteomic analysis of the response of Arabidopsis chloroplast proteins to high light stress. Proteomics. 2004;4:3560–3568.
- Zushi K, Ono M, Matsuzoe N. Light intensity modulates antioxidant systems in salt-stressed tomato (Solanum lycopersicum L. cv. Micro-Tom) fruits. Sci Hortic. 2014;165:384–391.
