Abstract
Maize is an important agricultural plant valued for its productivity and nutritive qualities. However, a deficiency of two essential amino acids (lysine and tryptophan) significantly reduces the nutritional quality of maize proteins. The recessive opaque-2 (o2) mutant has a greater content of lysine and tryptophan in their endosperm proteins and their bioavailability is better. Therefore, marker-assisted backcross breeding (MABB) was attempted to incorporate the o2 allele from the donor line VQL 1 into the genetic background of UMI 1200 to develop quality protein maize (QPM) lines. Foreground selection for the gene o2 was effected using tightly linked molecular marker umc1066, in UMI 1200 × VQL 1 backcross series. Further, background selection was done together with stringent phenotypic selection for agronomic traits, to accelerate recurrent parent phenome recovery. As a result, three advanced QPM maize lines (DBT 4-1-1/25-10/25-10/25-16, DBT 4-1-1/25-10/25-17/25-11 and DBT 4-1-1/25-10/25-17/25-13) carrying the opaque-2 allele in a homozygous state, similar to VQL 1 were developed. The lysine and tryptophan content of these lines ranged from 0.35% to 0.40% and 0.03 to 0.05%, respectively, which is on par with the VQL 1. Background analysis using 250 simple sequence repeats (SSRs) revealed up to 97% recurrent parent genome recovery. In conclusion, the newly developed QPM lines can be used in future maize breeding programmes to improve the nutritional traits.
Introduction
Maize (Zea mays L.) is a cereal crop which is cultivated widely throughout the world and has the highest production among all cereals. In India, maize is the third most important cereal crop after rice and wheat [Citation1]. Maize is grown throughout the year in all states of the country for human consumption and animal feed. India would require a double in production of maize to meet the domestic demand in next decade. Maize endosperm is rich in zein protein, which is alcohol soluble [Citation2,Citation3]. Zein is rich in glutamine (21–26%), leucine (20%), proline (10%) and alanine (10%), but deficient in important essential amino acids, for example, lysine (2.0% in the protein) and tryptophan (0.4% in the protein). The deficiency in these two amino acids essential for building proteins in humans leads to protein malnutrition [Citation4]. To solve the problem of malnutrition, a potential solution is sought in the adoption of quality protein maize (QPM) varieties possessing balanced protein due to higher lysine and tryptophan. Normal maize protein contains a lower level of lysine (0.16–0.26%) and tryptophan (0.02–0.06%) which is less than half of the recommended dose specified for human nutrition [Citation5,Citation6].
At the beginning, a major goal of breeding programmes was to convert the normal genotype to a QPM version through the conventional backcross approach [Citation7]. However, due to the linkage drag, several undesirable agronomic traits, such as soft endosperm and damaged kernels leading to severe susceptibility to pest and fungal attack, finally resulted in yield loss in the developed lines and, thus, to the non-acceptance of QPM maize [Citation8,Citation9]. Hence, converting the commercial maize lines into QPM cannot be achieved only by conventional backcross breeding. In this context, marker-assisted backcross breeding (MABB), as a form of marker-assisted selection (MAS), acts as an attractive tool to develop QPM maize. The MABB provides the information needed to reduce the number of generations of backcrossing, to combine (i.e. stack’) transgenes, ‘native’ genes or quantitative trait locus (QTL) into one inbred or hybrid line quickly, and to maximize the recovery of recurrent parent genome (RPG) in the backcross-derived progenies. Generic MABB schemes suitable for maize breeding programmes have been described in single genes [Citation9–13].
To date, many natural maize mutants conferring higher lysine and tryptophan content have been identified, that is opaque-2 (o2), floury-2 (fl2), opaque-7 (o7), opaque-6 (o6) and floury-3 (fl3) [Citation6]. Of these, the o2 mutation is considered to be the most suitable for genetic manipulation in breeding programmes aimed at developing maize with high lysine and tryptophan. A strong effort was taken by the plant breeders all over the world to transfer this opaque-2 gene into a high yielding commercial hybrid or cultivars. Gene o2 is located in chromosome 7 [Citation14] and the linked allele-specific simple sequence repeat (SSR) markers for the o2 gene are available for use in molecular breeding [Citation15,Citation16]. By introgression of the o2 gene into commercial hybrids or cultivars with MABB, the genetically modified o2 maize, also known as QPM, shows an improved lysine content of approximately 0.4% [Citation9]. Thus, the objectives of this research were to transfer opaque-2 into a maize inbred line using MABB and to evaluate the effect of this gene on lysine and tryptophan in advanced lines.
Materials and methods
Plant genetic materials
The present study aimed to introgress the o2 gene from VQL 1 into UMI 1200 through MABB. UMI 1200 (Female parent of TNAU maize Hybrid Co 6) is a well-adapted elite maize inbred line from Tamil Nadu used as a recurrent parent. It is characterized as dent grain type and the cob length is approximately 13.7 cm with 14 kernels per row and 100 grain weight of 25 g. UMI 1200 was obtained from Tamil Nadu Agricultural University, Coimbatore, India. VQL 1 carrying the o2 gene was used as a donor parent. It was obtained from Vivekananda Parvatiya Krishi Anusandhan Sansthan, Almora, India. All the field experiments were conducted at Agricultural Research Station (ARS), Tamil Nadu Agricultural University, Bhavanisagar, Tamil Nadu, India.
MABB strategy
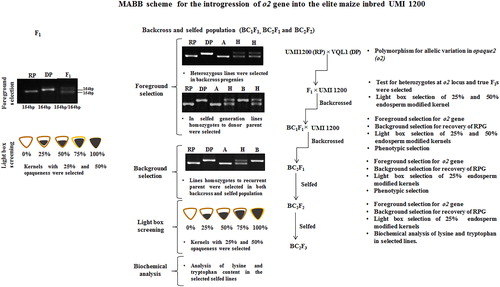
The MABB scheme that we used in developing improved lines is shown in . Crosses were made between elite maize inbred, UMI 1200 (recurrent) and VQL 1 (QPM donor) at Kharif 2015. The F1s were raised during Rabi 2016 and tested for heterozygosity using the o2 gene-specific SSR marker and the true F1s were backcrossed with the recurrent parent. BC1F1 progenies were grown during Kharif 2016 and were subjected to foreground selection using o2 gene-specific marker. Desirable plants with high recovery of the RPG and morphological similarity to the recurrent parent were further backcrossed to raise the BC2F1 population during Rabi 2017. The selected plants with high RPG and phenotypic similarity to the recurrent parent were selfed and a BC2F2 generation was produced. The BC2F2 population was raised during Kharif 2017 and selected plants were selfed to advance progenies and BC2F3 raised during Rabi 2018.
Figure 1. General scheme of marker-assisted backcross breeding (MABB) for the conversion of normal maize inbred line UMI 1200 into QPM version using the donor VQL 1. RP: recurrent parent (UMI 1200); DP: donor parent (VQL 1); A: homozygous allele to recurrent; B: homozygous allele to donor; H: heterozygous allele. Three levels of selection were carried out in developing the QPM version of UMI 1200: (i) foreground selection to select the plants with target allele o2; (ii) background selection to recover maximum RPG recovery; (iii) phenotypic selection for agronomic traits and endosperm modification.
Molecular marker assay
Genomic DNA isolation and PCR analysis
Genomic DNA was isolated from the leaves of 15-day-old seedlings using the standard cetyl trimethylammonium bromide (CTAB) method [Citation17]. Each DNA sample was diluted to a working concentration of 50 ng/μL for polymerase chain reaction (PCR) analysis. The PCR (Eppendorf, Hamburg, Germany) was carried out in 10-μL reaction mixtures containing 1 μL of 50 ng/μL genomic DNA as the template, 7 μL of 1× master mix RED with 1.5 mmol/L MgCl2 (Amplicon), 0.6 μL of the primer pairs (F and R) and 1.4 μL of Milli-Q water. The PCR cycle consisted of initial denaturation at 94˚C for 7 min, followed by 35 cycles of amplification at 94˚C for 30 s (denaturation), 60˚C for 30 s (annealing) and 72˚C for 45 s (extension). A final extension step at 72˚C for 7 min was followed by termination of the cycle at 4˚C. The resulting PCR amplicons of o2-linked marker were resolved in 3% agarose gel at 100 V for 3 h. The resolved amplified products were visualized using a gel documentation system (Bio-Rad, CA, USA).
Marker-assisted foreground selection
Three SSR markers (phi057, phi112 and umc1066) linked to the o2 locus were used for foreground selection. Parental polymorphism survey was conducted between the recurrent and donor parents. Of the three SSR markers for o2, umc1066 showed a distinct polymorphism between the recurrent and the donor parent. This marker was further used for foreground selection in backcrossed and selfed individuals. Chi-square (χ2) test was performed to test the goodness of fit at the o2 locus in each of the generations.
Marker-assisted background analysis
Parental polymorphism survey was conducted between the recurrent and donor parents using a panel of 250 microsatellite markers spanning uniformly across the maize genome. The primer sequences for 250 SSR markers were adapted from the maize genome database SSR marker resource (www.maizegdb.org) and custom synthesized (Eurofins, Bangalore, India). The resulted polymorphic SSRs spanning evenly across the genome were used to recover the RPG in BC1F1, BC2F1 and BC2F2 generations.
Endosperm modification
The endosperm modification of kernels can be observed through a light table apparatus. The o2o2 genotypes carry an undesirable character, namely, kernel softness and this can be seen as complete opaqueness when observed through a light table [Citation6]. The genes for endosperm hardness/softness segregate and hence varying levels of opaqueness are observed in the kernels when subjected to light box test. A kernel with the O2o2 or O2O2 genotype is normal maize, that is it does not have the softness and undesirable kernel characteristics associated with the o2o2 genotype and is therefore translucent; however, it shows some insignificant degree of opaqueness [Citation6]. Light table selection is an indirect measure by which the kernels with o2o2 genotype can be picked out based on the degree of opaqueness. The kernels were divided into five group, namely 0%, 25%, 50%, 75% and 100% opaqueness depending on the transparency of the kernels visible in the light table test. The kernels with 0% opaqueness should not be selected in the early generation, because they may have a modified o2o2 gene or heterozygous (O2o2) or homozygous dominant (O2O2) kernels. Kernels with 25% opaqueness almost surely are o2o2, whereas 50% seeds certainly have the o2o2 genotype [Citation6]. Although the seeds with 75% and 100% opaqueness have o2o2, selecting such types may result in very low activity of modifiers and acquire several undesirable characters such as kernel cracking, susceptibility to pest and diseases [Citation6,Citation8]. Hence in the present study, kernels with 25% and 50% opaqueness were selected to raise the BC1F1 population, whereas in the BC2F1 and BC2F2 generations, the kernels with 25% opaqueness were selected to forward next generation.
Morphological characterization
The original inbred line, F1 progenies, BC1F1, BC2F1 and BC2F2 progenies were characterized for eight morphological characters, namely days to tasselling (days), days to silking (days), plant height (cm), tassel length (cm), number of tassel branches, ear height (cm), leaf length (cm), leaf width (cm) and seven grain yield-related traits namely, cob length (cm), cob girth (cm), number of kernel rows per cob, number of kernels per row, cob weight (g), kernel type (dent/flint) and 100 grain weight (g).
Estimation of lysine and tryptophan content in endosperm
The foreground positive BC2F2 lines were selfed and selfed cobs were subjected to lysine and tryptophan estimation in according to the method of Galicia et al. [Citation18]. All the samples were replicated twice for the analysis. Two blanks and four checks (2 QPM and 2 normal) were included along with the sample. The absorbance of lysine and tryptophan was measured by a spectrophotometer V-770 UV-VIS-NIT (OK, USA) at 390 nm and 560 nm, respectively. The lysine and tryptophan contents were calculated as µg/µg as described by Liu et al. [Citation19].
Results and discussion
Marker-assisted introgression of o2 allele
Normal maize protein contains lower levels of lysine and tryptophan, which are less than half of the recommended dose specified for human nutrition [Citation6]. Enhancement of these amino acids has been achieved in maize by employing o2 mutant that led to the development of QPM. Introgressing of an o2 recessive allele through conventional backcross breeding involves 6–7 generations of backcrossing, and it is expensive, laborious and time-consuming due to large-scale phenotyping of the segregating population. Therefore, in the present investigation, MABB was successfully utilized for developing QPM version of UMI 1200. Among the three SSR markers which were linked to the o2 locus, umc1066 showed polymorphism between the recurrent and the donor parent. The nature of the polymorphism exhibited by umc1066 was co-dominant and hence it was easy to discriminate between homozygotes and heterozygotes. Identification of heterozygotes in the early stage before flowering facilitated the exclusion of plants with dominant homozygotes (O2/O2) in the backcross population. To develop a QPM version of UMI 1200 inbred, the F1s from the cross UMI 1200 ×VQL 1 were tested for the presence of QPM allele with the gene-specific SSR umc1066 and the selected plants were backcrossed to generate BC1F1. A total of 232 plants in BC1F1 and 218 plants in BC2F1 were subjected to foreground screening with umc1066 and this resulted in 130 and 112 heterozygotes, respectively, in each generation. The identified heterozygous plants were backcrossed to their recurrent parent to develop the next generation. The segregation pattern in these backcross populations reasonably fitted with the Mendelian segregation ratio of 1:1 (). Similar results are also obtained by other researchers [Citation4,Citation8,Citation19,Citation20]. Foreground screening of 158 BC2F2 plants led to the identification of 21 homozygous plants for recessive allele (o2/o2). However, the chi-square test showed that the population deviated from the expected Mendelian segregation pattern of 1:2:1 (), which is similar to the results of Jompuk et al. [Citation21]. This deviation may be due to the presence of many segregation distortion regions (SRDs) in the maize genome [Citation22]. Segregation distortion in maize was first reported by Mangelsdorf and Jones [Citation23]. There are several genes that cause segregation distortion and if such genes segregate in a population, then the markers close to them would exhibit distorted ratios [Citation24]. Other reasons may include small population size and the genetic background of the target allele. Comparable results were reported in a study by Muthusamy et al. [Citation25] in which they identified four out of seven crosses deviating from the expected ratio of 1:1 in their backcross population. Hossain et al. [Citation26] also recorded segregation distortion in their backcross and selfed population. Foreground screening of F1, BC1F1, BC2F1 and BC2F2 was presented in .
Figure 2. Foreground screening of F1, BC1F1, BC2F1 and BC2F2 individuals of UMI 1200 × VQL 1 using umc1066 primer. (A) Foreground selection in F1 individuals; lanes 1–16, F1 progenies. (B) Foreground selection in BC1F1 individuals; lanes 1–16, BC1 F1 progenies. (C) Foreground selection in BC2F1 individuals; lanes 1–17, BC1 F2 progenies. (D) Foreground selection in BC2F2 individuals; lanes 1–17, BC2 F2 progenies. M: 100 bp ladder; P1: UMI 1200; P2: VQL 1.
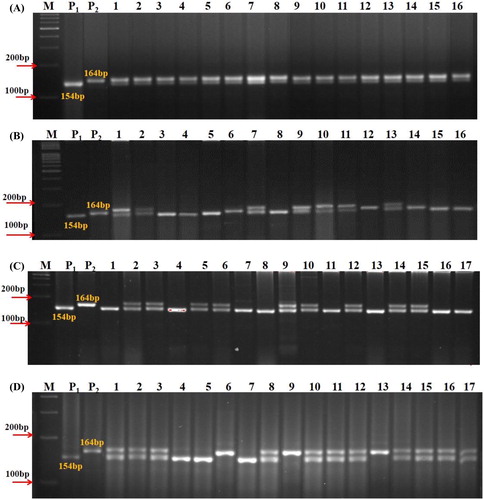
Table 1. Segregation pattern of the o2 allele in backcross and selfed generations of cross UMI 1200 × VQL 1.
A high recovery of RPG was achieved in the backcross progenies using 250 genome-wide SSRs distributed in all 10 chromosomes. Among the 250, only 106 markers revealed polymorphism between the recurrent and the donor parent. These 106 markers were found to be dispersed in all the 10 chromosomes of maize which varies from 8 to 11 markers per chromosome. These polymorphic markers were used for screening the foreground positive individuals of BC1F1, BC2F1 and BC2F2. The individuals that were homozygous to the recurrent parent were selected and forwarded to the subsequent generation to recover the RPG. The recovery per cent of BC1F1s ranged from 62.1% to 74.2% and seven plants (with >70.7% RPG) were selected and advanced to BC2F1. A recovery range of 82.4–92.8% was observed in the BC2F1 population and two individuals with more 90% recovery () were advanced to BC2F2s. In the BC2F2 population, the recovery range was between 87.4% and 96.6% and the three plants with highest recovery (DBT 4-1-1/25-10/25-10/25-16 with 96.6% RPG, DBT 4-1-1/25-10/25-17/25-11 with 92.4% and DBT 4-1-1/25-10/25-17/25-13 with 95.4%) were selfed to generate BC2F3. In this study, a rapid recovery of 90% RPG was achieved in three generations. This was made possible with MAS. However, in a conventional backcross approach, 5–6 backcross generations are needed to attain 90% recovery. In addition, o2 is a recessive gene and after successful generation of each backcross population, selfing has to be done to reject the homozygous dominant and heterozygous individuals [Citation27,Citation28]. This would require another four generations consuming a total of 7–8 years to develop a line. To overcome this time consumption, several researchers have utilized MAS strategy in developing QPM inbreds and hybrids. Marker-assisted foreground selection helped in selecting the individuals with the target gene and the background selection facilitated selecting the individuals with high RPG recovery [Citation8,Citation9,Citation29]. Our study also proved the advantage of MABB in reducing the time period for introgression of the target gene into an elite inbred line. The three selected individuals of BC2F2 showed morphological similarity with the recurrent parent, desirable endosperm modification of 25% opaqueness and the kernel dent, which is a predominant character of UMI 1200. The banding patterns of the selected BC1F1 and BC2F2 individuals are presented in .
Figure 3. Background selection in selected individuals. Amplification with: umc1710 in BC2F1 (A); umc2101 BC1F1 (B); umc1026 in BC2F2 (C); umc1060 in BC2F2 (D); umc1924 in BC2F2 (E); umc1827 in BC2F2 (F); umc2143 in BC2F2 (G); umc1857 in BC2F2 (H); umc1185 in BC2F2 (I); nc012 in BC2F2 (J). M: 100 bp ladder; P1: UMI 1200; P2: VQL 1; Lane Citation1: DBT4-1-/25-10/25-10/25-16; Lane Citation2: DBT4-1-/25-10/25-17/25-11; Lane Citation3: DBT4-1-/25-10/25-17/25-13. The plant was scored AA if it contains the UMI 1200 allele, as BB if it contains the VQL 1 allele and as AB if it contains both UMI 1200 allele and VQL 1 allele (i.e. heterozygotes).
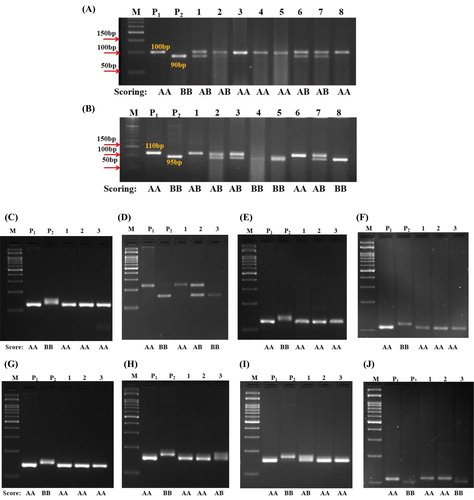
Table 2. Recovery percentage of recurrent parent genome (RPG) in backcross populations of cross UMI 1200 × VQL 1.
Endosperm modification in the introgressed progenies
Although we selected plants with o2-specific foreground markers, light box screening is required in every generation to select kernels with 25% and 50% and to exclude kernels with 0%, 75% and 100% as mentioned earlier. Previously, Hossain et al. [Citation26] selected seeds with 25% and 50% opaqueness to generate hybrids. Similarly, in this study, in F1 and BC1F1 generations, seeds with 25% and 50% modification were forwarded. From BC2F1 onwards, the kernels with 25% modification were advanced to the next generation. In the harvested cobs of BC2F2 (in plants that were o2o2 homozygotes), the kernels expressed only 0% and 25% modification () proving that the population had obtained the homozygosity for endosperm modification.
Agronomic performance of MAS derived lines
The morphological characters such as grain yield and its attributes are recovered rapidly through MAS [Citation25]. In the present study, the BC2F2 lines showed a wide range of segregation in the field. However, the three foreground positive lines (homozygous for the target allele, o2o2) namely DBT 4-1-1/25-10/25-10/25-16, DBT 4-1-1/25-10/25-17/25-11 and DBT 4-1-1/25-10/25-17/25-13 were found to have high degree of resemblance to the recurrent parent (). The similarity percentage of line DBT 4-1-1/25-10/25-10/25-16 ranged from 85.71% (number of kernel rows per cob) to 96.77% (days to 50% silking). The lines DBT 4-1-1/25-10/25-17/25-11 and DBT 4-1-1/25-10/25-17/25-13 exhibited more than 80% resemblance with the recurrent parent for all the observed traits except number of kernel rows per cob (71.43%) and cob girth (78.43%), respectively (). Related studies by Gupta et al. [Citation30] also showed that the plant and cob characters of Vivek QPM 9 were similar to those of VMH 9 (non-QPM) developed. Hossain et al. [Citation26] developed three MAS-derived inbreds and hybrids that showed high degree of resemblance with their respective non-QPM inbreds and hybrid.
Table 3. Recovery percentage for morphological traits of BC2F2 progenies in comparison with recurrent and donor parents.
Lysine and tryptophan levels in the endosperm of MAS-derived lines
The presence of the o2 allele in recessive condition reduces the level of α-zeins, thereby increasing the level of lysine and tryptophan [Citation20,Citation30,Citation31]. The lysine and tryptophan concentrations were analyzed for VQL 1 (donor), UMI 1200 (recurrent) and the three selected lines (DBT 4-1-1/25-10/25-10/25-16, DBT 4-1-1/25-10/25-17/25-11 and DBT 4-1-1/25-10/25-17/25-13) using the calorimetric method [Citation18]. The lysine and tryptophan concentrations were given per unit of sample and were expressed as per cent (it is not the per cent of lysine/tryptophan in protein). The results showed that the lysine content for the VQL 1 and UMI 1200 was 0.42% and 0.26%, respectively, whereas the tryptophan concentrations were 0.05 and 0.013%, respectively. The lysine concentration in selfed cobs of the selected BC2F2 lines varied from 0.35% to 0.40%, whereas the tryptophan content varied from 0.03 to 0.05%, which indicates a twofold increase in the level of lysine and tryptophan as compared to the recurrent parent. Liu et al. [Citation19] in their study reported a mean concentration of 0.30% lysine and 0.85% tryptophan in a BC2F3 population.
Conclusions
This study demonstrated the successful introgression of the o2 gene from VQL 1 and restoration of the UMI 1200 genome through MABB. As a result, a set of three high lysine and tryptophan lines was developed. These advanced lines can be used in future breeding programmes, such as pyramiding with other varieties, to improve the nutritional traits, such as lysine and tryptophan.
Acknowledgements
The authors thank the Centre of Innovation, Agricultural College and Research Institute, Tamil Nadu Agricultural University, Madurai, for providing lab facilities.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was financially supported by grants from the Department of Biotechnology, Government of India through the project entitled ‘Enrichment of nutritional quality in maize through molecular breeding’ (BT/PR10922AGII/106/9442014 dt.25.3.2015). The funders had no role in the work design, data collection and analysis, or decision and preparation of the manuscript.
References
- Krishna MS, Reddy SS, Satyanarayana SD. Marker-assisted breeding for introgression of opaque-2 allele into elite maize inbred line BML. Biotech. 2017;7( 3):165.
- Shewry PR, Tatham AS. The prolamin storage proteins of cereal seeds: structure and evolution. Biochem J. 1990;267:1–12.
- Prasanna BM, Vasal SK, Kassahun B. Quality protein maize. Curr Sci. 2001;81:1308–1319.
- Tripathy SK, Ithape DM, Maharana M, et al. Quality protein maize (QPM): genetic basis and breeding perspective. Trop Plant Res. 2017;4:145–152.
- Bjarnason M, Vasal SK. Breeding of quality protein maize (QPM). Plant Breed Rev. 1992;9:181–216.
- Vivek BS, Krivanek AF, Palacios-Rojas N. Breeding quality protein maize (QPM): protocols for developing QPM cultivars. Mexico: CIMMYT; 2008.
- Glover DV, Mertz ET. Corn. In: Olson RA, Frey KJ, editors. Nutritional security of cereal grains: genetic and agronomic improvement. Madison (WL): Agron Monogr 28 ASA CSSA and SSSA; 1987.
- Babu R, Nair SK, Kumar A, et al. Two-generation marker-aided backcrossing for rapid conversion of normal maize lines to quality protein maize (QPM)). Theor Appl Genet. 2005;111:888–897.
- Hospital F, Chevalet C, Mulsant P. Using markers in gene introgression breeding programs. Genetics. 1992;132:1199–1210.
- Ragot M, Biasiolli M, Delbut MF, et al. Marker-assisted backcrossing: a practical example. In: Berville A, Tersac M, editors. Les Colloques, no. 72, techniques et utilisations des marqueurs moleculaires. Paris (France): INRA; 1995. p. 45–56.
- Frisch M, Melchinger AE. Marker assisted backcrossing for introgression of a recessive gene. Crop Sci. 2001;41:1485–1494.
- Hospital F, Charcosset A. Marker-assisted introgression of quantitative trait loci. Genetics. 1997;147:1469–1485.
- Bouchez A, Hospital F, Causse M, et al. Marker-assisted introgression of favorable alleles at quantitative trait loci between maize elite lines. Genetics. 2002;162:1945–1959.
- Gibbon BC, Larkins BA. Molecular genetic approaches to developing quality protein maize. Trends Genet. 2005;21:227–233.
- Bantte K, Prasanna BM. Simple sequence repeat polymorphism in quality protein maize (QPM) lines. Euphytica. 2003;129:337–344.
- Yang W, Zheng Y, Ni S, et al. Recessive allelic variations of three microsatellite sites within the O2 gene in maize. Plant Mol Biol Rep. 2004;22:361–374.
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325.
- Galicia L, Nurit E, Rosales A, et al. Maize nutrition quality and plant tissue analysis laboratory. Mexico: CIMMYT; 2008. p. 10–17.
- Liu L, Jeffers D, Zhang Y, et al. Introgression of crtRB1 gene into quality protein maize inbred line using molecular markers. Mol Breed. 2015;35:154.
- Gupta HS, Babu R, Agrawal PK, et al. Accelerated development of quality protein maize hybrid through marker-assisted introgression of opaque-2 allele. Plant Breed. 2013;132:77–82.
- Jompuk C, Cheuchart P, Jompuk P, et al. Improved tryptophan content in maize with opaque-2 gene using marker assisted selection (MAS) in backcross and selfing generations. Kasetsart J. (Nat Sci) 2011;45:666–674.
- Lu H, Romero-Severson J, Bernardo R. Chromosomal regions associated with segregation distortion in maize. Theor Appl Genet. 2002;105:622–628.
- Mangelsdorf PC, Jones DF. The expression of Mendelian factors in the gametophyte of maize. Genetics. 1926;11:423–455.
- Zamir D, Tadmor Y. Unequal segregation of nuclear genes in plants. Botanical Gazette. 1986;147:355–358.
- Muthusamy V, Hossain F, Thirunavukkarasu N, et al. Development of β-carotene rich maize hybrids through marker assisted introgression of β-carotene hydroxylase allele. PLoS One. 2014;9:e113583.
- Hossain F, Vignesh M, Pandey N, et al. Marker-assisted introgression of opaque 2 allele for rapid conversion of elite hybrids into quality protein maize. J Genet. 2018;97:287–298.
- Singh BD. Principles of plant breeding. New Delhi: Kalyani Publishers; 2012.
- Chen HM, Zhang YD, Chen W, et al. Improving grain yield and yield components via backcross procedure. Maydica. 2010;55:145–153.
- Tanksley SD. Molecular markers in plant breeding. Plant Mol Biol Rep. 1983;1:3–8.
- Gupta HS, Agrawal PK, Mahajan V, et al. Quality protein maize for nutritional security: rapid development of short duration hybrids through molecular marker-assisted breeding. Curr Sci. 2009;96:230–237.
- Moro GL, Habben JE, Hamaker BR, et al. Characterization of the variability in lysine content for normal and opaque2 maize endosperm. Crop Sci. 1996;36:1651–1659.

