Abstract
We studied the petal number trait in a population obtained after self-pollination of R. x damascena f. trigintipetala following analysis of molecular markers which have previously been mapped near the major dominant locus Blfo/d6 determining this trait in other rose species including R. multiflora and R. hybrida. The results showed that the same genetic mechanism, which determines the petal number trait in R. multiflora and R. hybrida also controls the trait in R. x damascena f. trigintipetala and is related to the dominant effect of a single copy allele in the tetraploid genome of this species. We also analyzed the expression of several flower homeotic genes including R. x damascena APETALA1/FUL-like (paleo AP1 type), R. x damascena euAPETALA 3 (euAP3 line) and R. x damascena AGAMOUS in early stage flower buds corresponding to plants with double and simple flowers. The obtained results showed that only R. x damascena AGAMOUS was differentially expressed between the samples of double and simple flowers, its relative expression being upregulated 3.5-fold in simple flowers. We further cloned and sequenced the four genomic clones of R. x damascena AGAMOUS and studied the potential additive effect of this gene by analysing the segregation of its four alleles in the population of self-pollinated R. x damascena. Analysis of variance of the data for petal number and allele segregation did not show a statistically significant effect of any allele configuration of the AGAMOUS gene on the petal number trait in R. x damascena f. trigintipetala.
Introduction
Although more than 200 species have been identified in the genus Rosa, only a few of them have been industrially grown for production of rose essential oil products, which are widely applied as base components in perfumery and cosmetics products by the biggest producers in the market [Citation1]. Rosa x damascena Mill, also known as the oil-bearing rose, has been grown for centuries for its highly scented flowers containing high amounts of essential oil components. The production of rose oil as well as other aromatic products including rose water, rose concrete and rose absolute from the flowers of R. x damascena is an important traditional industry in a number of countries in Europe, the Middle East and Northern Africa including Bulgaria, Turkey, Iran, India, Pakistan, Morocco and several others [Citation2]. Although other rose species like R. gallica and R. centifolia have also been grown for rose oil production during the years, currently the predominant rose species grown worldwide for industrial production of rose essential oil products is R. x damascena Mill.
The main product obtained from the flowers of R. x damascena is the essential oil produced by water–steam distillation of its flowers. The two biggest producers of rose oil supplying more than 90% of the rose oil to the world market are Bulgaria and Turkey [Citation3]. Several studies have shown that the entire industrial cultivation of R. x damascena in these two countries is based on a single genotype of this rose species, namely the f. trigintipetala form, which has 30–35 petals in its flowers and has been vegetatively propagated for centuries from a common ancestor [Citation4–6]. In contrast, a number of other genotypes of R. x damascena exist in Iran and other neighbouring countries, as today Iran and its region is considered to be the geographic origin of this species [Citation7,Citation8]. These studies, as well as the general trend towards diversification of natural products nowadays, clearly demonstrate that it is necessary to expand the local genetic resources of R. x damascena in the two countries that are the main producers of rose oil in the world. Several reviews have discussed the prospects of improving the oil rose and expanding its genetic resources in Bulgaria and Turkey through breeding based on cross-pollination involving intra- or inter-specific hybridization strategies [Citation9,Citation10]. A key element when implementing such strategies would be to maintain or increase the number of flower petals, whose epidermal layer is the main source of essential oil compounds [Citation11]. The number of petals in the diploid R. multiflora has been demonstrated to be a monogenic trait controlled by a major dominant allele of the Blfo/d6 locus mapped on LG3 in the unified genetic map of diploid roses [Citation12,Citation13]. Later studies also mapped the same locus in tetraploid roses based on QTL analysis [Citation14]. Additive effects of other loci in the rose genome are expected to further contribute to the petal number trait [Citation15].
Rosa x damascena is a tetraploid rose species with a possible triparental origin [Citation16]. Due to its complex genome and the hybrid nature of this species, genetic studies related to segregation of traits in populations developed with R. x damascena as a parent have not been implemented so far. In a recent study, Baydar et al. [Citation17] analysed a population obtained after self-pollination of R. x damascena f. trigintipetala in Turkey and observed large segregation of the number of petals in the population ranging from 5 to 114. In the present study, we analysed the segregation of the petal number trait and molecular markers located in the vicinity of the Blfo/d6 locus in a population of roses obtained after self-pollination of R. x damascena f. trigintipetala. We further analysed the expression of several flower homeotic genes in simple and double flower plants during the early stages of flower bud development and analysed the potential additive effect of the class C flower homeotic gene R. x damascena AGAMOUS.
Material and methods
Plant material and genomic DNA purification
Fully developed flowers of R. x damascena Mill f. trigintipetala and 143 plants derived from self-pollination of R. x damascena f. trigintipetala corresponding to phase 6 according to Rusanov et al. [Citation18] were picked in the early morning hours in June 2017, and were immediately frozen in liquid nitrogen for subsequent purification of genomic DNA. The frozen flowers were stored at −80 °C until they were milled into fine powder using a TissueLyser II (Qiagen) Laboratory Mill. Genomic DNA was purified using a GeneJET Plant Genomic DNA purification kit (Thermo Scientific, Waltham, MA) according to the manufacturer’s instructions. The concentration of the resulting genomic DNA was determined using Nanodrop 2000 (Thermo Scientific).
Morphological characterization of rose flowers
At least three fully developed flowers corresponding to phase 6 according to Rusanov et al. [Citation18] were collected from each of the analysed rose plants in the early morning hours in June 2017 and used for analysis of flower morphology. The parameters measured included flower diameter, number of petals and number of stamens. Flowers with 6 ± 1 petals were scored as simple flowers, flowers with petals >7 ≤ 20 were scored as semi-double flowers and flowers with petals >20 were scored as double flowers.
qPCR analysis of flower homeotic genes
Flower buds at the early stage of flower development corresponding to stage 1–4 according to Dubois et al. [Citation19] were used for total RNA purification. The buds were sampled from a plant possessing double flowers (30–35 petals) corresponding to the original genotype of R. x damascena f. trigintipetala and from a plant with simple flowers (six petals) obtained after self-pollination of R. x damascena f. trigintipetala. The flower buds were collected on 10 May 2017, prior to the start of the flowering season, and were immediately frozen in liquid nitrogen prior to purification of total RNA. Total RNA was purified using a CloneJET Plant RNA Purification Mini Kit (Thermo Scientific) according to the manufacturer’s instructions. Genomic DNA residues were removed using DNase I (Thermo Scientific) according to the manufacturer’s instructions. The concentration of the resulting total RNA was determined using Nanodrop 2000 (Thermo Scientific). SensiFAST™ SYBR Hi-ROX One-Step Kit (Thermo Scientific) was used to perform qPCR analysis using 40 ng of total RNA. Two genes with proven stable expression in R. hybrida encoding Translational Controlled Tumor Protein (TCTP) and Elongation Factor 1-alpha (EF1-alpha) () were used for signal normalization. The primer pairs used and the homoeotic genes analyzed are presented in . The online version of Primer 3 (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) was used for design of primers for R. x damascena euAPETALA3 (euAP3 line) and R. x damascena APETALA1/FUL. The assay was performed on an ABI 7300 Real-time PCR system (Applied Biosystems) using the following temperature programme: reverse transcription at 45 °C for 10 min, activation of the polymerase at 95 °C for 2 min; 40 cycles of denaturation at 95 °C for 5 s, primer hybridization/elongation at 60 °C for 27 s (fluorescence reading); dissociation step for analysis of melting curve including 95 °C for 15 s, 60 °C for 1 min, 95 °C for 15 s. Biorad Gene Expression Macro™ Version 1.1 software was used to determine the relative expression of the analysed genes.
Table 1. Primers used for analysis of expression of flower homeotic genes in early stages of flower development in Rosa x damascena by qPCR.
Cloning of the allelic forms of R. x damascena AGAMOUS
Cloning of the allelic forms of R. x damascena AGAMOUS, an ortholog of R. rugosa MASAKO C1, was done using a forward (5′-TGGACTCTGATGCCCAAAGA-3') and reverse primer (5′-GAAGGGAAATCTGGTCATGGC-3′) constructed using the online version of the Primer 3 software and GenBank deposited sequences of cDNAs from R. rugosa and R. hybrida. The constructed primer pair was used for PCR amplification of genomic DNA from R. x damascena f. trigintipetala. PCR reactions were performed on an Applied Biosystems 9700 PCR apparatus in a volume of 30 μL using 40 ng genomic DNA, 20 pmol right and reverse primer, and Phusion High-Fidelity PCR Kit (Thermo Scientific) according to the manufacturer’s conditions using the following temperature programme: denaturation at 98 °C for 30 s; 35 cycles of denaturation at 98 °C for 10 s, hybridization at 62 °C for 30 s, elongation at 72 °C for 4 min and a final elongation step at 72 °C for 10 min. The resulting PCR products were separated in an 0.8% agarose gel prepared with Top Vision Agarose (Thermo Scientific) visualized using Gelred (Biotium) and the approximate size determined using O’GeneRuler 1 kb DNA ladder (Thermo Scientific). The PCR products were purified from the gel using GeneJET Gel Extraction Kit (Thermo Scientific). The resulting purified PCR fragments were used for cloning in pJET1.2/blunt vector using a CloneJET PCR cloning kit (Thermo Scientific) and the obtained recombinant plasmids were transformed into competent Escherichia coli Dh5α cells by heat shock [Citation20]. Plasmid vectors from individual colonies were used for sequencing using pJET Fw (5′-CGACTCACTATAGGGAGAGCGGC-3′), pJET Rev (5′-AAGAACATCGATTTTCCATGGCAG-3′) as well as internal primers developed for primer walking. Sequencing was performed by Macrogen Inc. (Seoul, Republic of Korea) The four identified allelic forms were deposited in Genebank under the following accession numbers: MH593872, MH593873, MH593874 and MH593875.
Analysis of allelic configurations of R. x damascena mill AGAMOUS
The fluorescently labelled primers for amplification of allele-specific PCR products were designed using the online version of the Primer 3 software based on the sequences of the four identified alleles of R. x damascena AGAMOUS. The constructed primer pair consisted of the forward primer AGM2F (FAM-5′-TCAATAAACTGTGGCAGTGAAC-3′), fluorescently labelled at the 5′-end with FAM and the reverse primer AGM2R (5′-TTTGCTTGGTCATTGATCCT-3′). The temperature programme used for PCR amplification was as follows: denaturation at 95 °C for 3 min; 35 cycles of denaturation at 95 °C for 15 s, hybridization of the primers at 53 °C for 15 s, elongation at 72 °C for 30 s and a final elongation step at 72 °C for 5 min. Analysis of the amplified fragments was performed on an ABI 3130 (Thermo) automated sequencer using capillaries with a length of 36 cm, POP7 polymer and GeneScan 600 LIZ dye Size Standard (Thermo Scientific). Sizing of the PCR fragments was performed using the GeneMapper 4.0 software (Applied Biosystems, Foster City, CA).
Analysis of microsatellite loci
The PCR reactions were performed in a volume of 20 μL, comprising 40 ng genomic DNA, 5 pmol locus specific forward primer containing a 14-bp M13 tail at the 5′-end (), 20 pmol locus specific reverse primer (), 20 pmol forward M13 primer labelled at the 5′-end with FAM (FAM-5′-GTAAAACGACGGCCAGT-3′) used for fluorescent labelling of the products and 2× PCR mix MyTaq HS Mix, 2× (Bioline USA Inc.). The temperature programme included: denaturation at 95 °C for 3 min, 30 cycles of denaturation at 95 °C for 15 s, primer annealing at the temperatures shown in for 15 s, elongation at 72 °C for 30 s followed by 10 cycles of denaturation at 95 °C for 15 s, hybridization at 53 °C for 15 s, elongation at 72 °C for 30 s and a final elongation step at 72 °C for 10 min. Fragment analysis was performed on an ABI 3130 (Thermo) automated sequencer using capillaries with a length of 36 cm, POP7 polymer (Thermo) and GeneScan 600 LIZ dye Size Standard (Thermo). Sizing of the PCR fragments was performed using the GeneMapper 4.0 software (Applied Biosystems) and the allele lengths of the analysed loci were calculated after subtracting 17 bp corresponding to the length of the M13 primer used for fluorescent labelling.
Table 2. Primers used for analysis of microsatellite loci.
Table 3. Flower biometric analysisTable Footnote* of plants obtained after self-pollination of Rosa x damascena f. trigintipetala.
Statistical analysis and graphical presentation of data
Correlation analysis as well as analysis of variance (ANOVA) was performed using SPSS v. 21 (IBM SPSS Statistics, Armonk, NY). Microsoft Office 2016 was used for building box and whiskers plots.
Results and discussion
Flower morphology
A total of 143 plants from a population obtained after self-pollination of R. x damascena f. trigintipetala, were used for characterization of flower morphology parameters. Significant variation in the population was observed for all three measured parameters including flower diameter, number of petals and number of stamens (, ). Forty plants (28%) were identified as possessing simple flowers (6 ± 1 petals), 9 plants (6%) with semi-double flowers (>7 ≤ 20 petals) and 95 plants (66%) with double flowers (>20 petals). Correlation analysis showed statistically significant (p<.01) strong negative correlation (r= −0.785) between the number of petals and the number of stamens in the flowers. There was weak negative correlation between the number of petals and flower diameter (r= −0.242, p<.01) and weak positive correlation between the number of stamens and flower diameter (r = 0.269, p<.01). Roses with simple flowers were characterized by a large number of stamens, 110 ± 12, with a maximum number of 132. At the same time, significant variation in the number of both petals and stamens with mean values of 43 ± 20 and 70 ± 28, respectively was observed in plants with double flowers. The variation of petal number ranged from 21 to 103, while the number of stamens ranged from 0 to a maximum of 134, which was even higher compared to simple flowers.
Segregation of microsatellite markers located near the Blfo/d6 locus
Five microsatellite loci (Rh58, RhI402, Rh50, Rh65 and Rw11E5) derived from R. hybrida and R. wichuraiana and located in the region near the Blfo/d6 locus on LG3 in the unified genetic map of diploid roses were analysed in the original genotype of R. x damascena f. trigintipetala. Of the five tested markers, only Rh58, RhI402 and Rh65 produced clear and reproducible PCR products with allele sizes of 222, 228, 248 and 260 bp for Rh58, 194, 200 and 206 bp for RhI402 and 128 bp for Rh65. Since only locus Rh58 had four different alleles in the original genotype of the tetraploid R. x damascena, this made it the most suitable candidate for analysis of allele segregation in the population. shows the data distribution for number of petals among the plants in the population for each observed allele phenotype of Rh58 in a population of 143 plants obtained after self-pollination of R. x damascena f. trigintipetala. ANOVA analysis showed that the observed allele phenotype of the analysed locus had a statistically significant effect on the number of flower petals (p<.01). The analysis of the results presented in showed a well-expressed relation between certain allele phenotypes of the Rh58 locus and the formation of double (>20 petals) or simple flowers (6 ± 1 petals). In 92% of the cases, the presence of the A allele (222 bp) was associated with the development of double or semi-double flowers, while in 81% of the cases its absence was associated with the development of simple flowers.
Figure 2. Box-and-whiskers plot showing the distribution of data for number of petals among plants with different allele phenotype of the microsatellite marker Rh58 in plants from a segregating population obtained after self-pollination of Rosa x damascena f. trigintipetala. A, B, C and D designate the scored allele sizes (A = 222 bp, B = 228 bp, C = 248 bp and D = 260 bp).
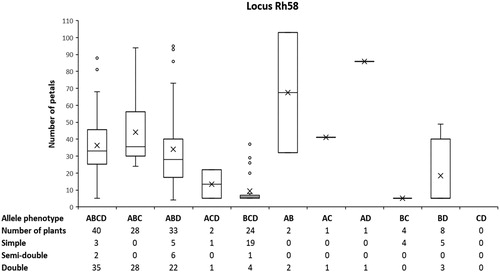
The obtained results show that the petal number trait in the tetraploid R. x damascena is controlled by the presence of a single copy of a dominant allele of the major locus designated as Blfo/d6 on LG3 in the genetic linkage map of roses [Citation12,Citation13]. Until recently, specific genes associated with the major Blfo/d6 locus had not been identified and isolated and its mapping in the R. multiflora genetic map was done following the segregation of the morphological trait as a monogenic trait and its linkage with certain molecular markers. Recently, the complete genome sequence of R. chinensis was published, the authors also pointing out APETALA2/TOE as a candidate gene located in the region of the major locus controlling the petal number trait [Citation21]. According to the ABC flower model, AP2 negatively regulates the expression of AGAMOUS [Citation22]. However, further confirmation and verification of the role of APETALA2/TOE will be needed, which would then allow the use of gene-specific markers in breeding programmes without a compromise of potential recombination between the used marker and the locus determining the trait of interest.
qPCR and segregation analysis of R. x damascena AGAMOUS
The observed large variation in the group of double flower roses in the segregating population suggests the influence of other loci in addition to the major locus controlling the trait. The expression of R. x damascena AGAMOUS and two other flower homeotic genes including R. x damascena APETALA1/FUL-like (paleo AP1 type) and R. x damascena euAPETALA 3 (euAP3 line) were studied during the early stages of flower development (stages 1–4 according to Dubois et al.[Citation19]) in flower buds of the original genotype of R. x damascena f. trigintipetala possessing double flowers (30–35 petals) and in a plant obtained after self-pollination of R. x damascena f. trigintipetala characterized by simple flowers (6 ± 1 petals). Of the three genes tested, only R. x damascena AGAMOUS showed statistically significant difference in expression between the analysed samples (): it was 3.4-fold upregulated in buds corresponding to simple flowers compared to double flower buds. Although the ortholog of A. thaliana AGAMOUS in roses has not been shown to be located in QTL loci controlling the petal number trait in roses and its location in the rose genetic map is on a different linkage group compared to the major Blfo/d6 locus, its expression has a direct influence on the petal number trait and the transition of stamens to petals in double flowers [Citation19]. We cloned and sequenced the four alleles of R. x damascena AGAMOUS and analysed their segregation in a population of 143 plants obtained after self-pollination of R. x damascena f. trigintipetala. shows the distribution of the data for petal number compared to the observed allele phenotype of R. x damascena AGAMOUS in the population. As seen from , there was no direct relation between the number of petals and allele phenotype for the majority of allele phenotypes. Although some allele phenotypes like the one designated as ABD in showed increased number of plants with double flowers, ANOVA analysis showed that the allelic configurations of R. x damascena AGAMOUS had no statistically significant influence on the number of petals (at p<.05). Additional ANOVA analysis after filtering out plants with simple flowers also showed no significant influence of the allelic configurations of R. x damascena AGAMOUS on the number of petals, thus suggesting that the gene is not likely to have an additive effect on the petal number trait in R. x damascena.
Figure 3. Expression level of Rosa x damascena AGAMOUS (RdAG), R. x damascena APETALA1/FUL-like (RdAP1/FUL-like) and R. x damascena euAPETALA3 (RdAP3). RdD, early stage flower buds of R. x damascena f. trigintipetala with double flowers. RdS, early stage flower buds of a simple flower plant (6 ± 1 petals) obtained by self-pollination of R. x damascena f. trigintipetala.
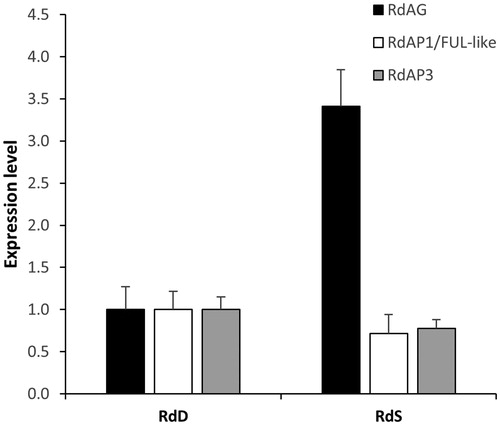
Figure 4. Box-and-whiskers plot showing the distribution of data for number of petals among plants with different allele phenotypes of Rosa x damascena AGAMOUS in a segregating population of 143 plants obtained by self-pollination of R. x damascena f. trigintipetala. A, B, C and D designate the 4 different alleles of R. x damascena AGAMOUS scored by using the AGM2 primer pair (A = 147 bp, B = 153 bp, C = 155 bp and D = 163 bp).
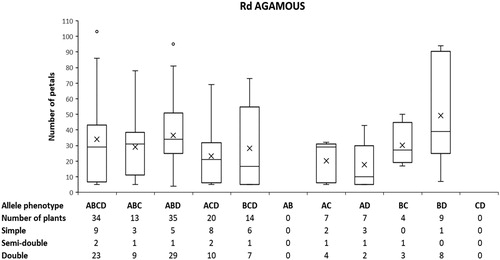
Our results related to the expression of the class C flower homeotic gene AGAMOUS are also consistent with the A. thaliana ABC model of flower development [Citation22,Citation23] and with the results of studies in R. hybrida, R. gallica, R. chinensis and R. rugosa [Citation19], according to which the expression of the homoeotic gene from Class C AGAMOUS is directly related to the change in the number of petals, and its increased expression in the early stages of flower bud development is associated with a decrease in their number at the expense of the number of stamens. The analysis of the segregation of its four different alleles in the population of self-pollinated R. x damascena did not appear to statistically influence the trait or to infer an additive effect. This supports the hypothesis that the factor which determines the double flower phenotype is either the APETALA2/TOE gene itself or one located very close to it [Citation21]. Analysis of other genes linked to QTL loci controlling the double flower trait will be necessary in order to obtain a set of molecular markers for precise control of the trait in breeding programmes in R. x damascena.
Conclusions
The results obtained in this study clearly showed that the double flower trait in the tetraploid R. x damascena is controlled by a single copy dominant allele of the locus Blfo/d6 previously mapped in the diploid R. multiflora. The results also indicated that the genetic background which determines the number of petals in the flowers of the tetraploid R. x damascena f. trigintipetala flowers is similar to that operating in the diploid R. multiflora and the teraploid R. hybrida. The study did not show the petal number of R. x damascena progeny plants to be influenced significantly by the allelic combination of the Class C flower homeotic gene AGAMOUS. The results from the present study offer direct application of molecular markers linked to the Blfo/d6 locus in breeding programmes for improvement of the industrially cultivated R. x damascena and efficient early selection of desired double flower plants.
Acknowledgements
The authors would like to thank Marina Alexeeva (Master degree student at the Sofia University) and Rumyana Velcheva (technician at ABI) for the excellent technical assistance.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Smulders M, Arens P, Koning-Boucoiran C. Rosa. Wild crop relatives: genomic and breeding resources. Berlin, Heidelberg: Springer; 2011. p. 243–275.
- Rusanov K, Kovacheva N, Atanassov A, et al. Rosa damascena Mill., the oil-bearing Damask rose: genetic resources, diversity and perspectives for molecular breeding. Floriculture Ornamental Biotech. 2009;3:14–20.
- Kovacheva N, Rusanov K, Atanassov I. Industrial cultivation of oil bearing rose and rose oil production in Bulgaria during 21st century, directions and challenges. Biotechnol Biotechnol Equip. 2010;24:1793–1798.
- Agaoglu Y, Ergul A, Baydar N. Molecular analyses of genetic diversity of oil rose (Rosa damascena Mill.) grown in Isparta (Turkey) region. Biotechnol Biotechnol Equip. 2000;14:16–18.
- Baydar NG, Baydar H, Debener T. Analysis of genetic relationships among Rosa damascena plants grown in Turkey by using AFLP and microsatellite markers. J Biotechnol. 2004;111:263–267.
- Rusanov K, Kovacheva N, Vosman B, et al. Microsatellite analysis of Rosa damascena Mill. accessions reveals genetic similarity between genotypes used for rose oil production and old Damask rose varieties. Theor Appl Genet. 2005;111:804–809.
- Tabaei-Aghdaei SR, Hosseini Monfared H, Fahimi H, et al. Genetic variation analysis of different populations of Rosa damascena in NW. Iran using RAPD markers. Iran J Bot. 2006;12:121–127.
- Kiani M, Zamani Z, Khalighi A, et al. Microsatellite analysis of Iranian Damask rose (Rosa damascena Mill.) germplasm. Plant Breeding. 2010;129:551–557.
- Rusanov K, Kovacheva N, Atanassov A, et al. Rosa damascena Mill., the oil-bearing Damask rose: genetic resources, diversity and perspectives for molecular breeding. Floriculture Ornamental Biotechnol. 2009;3:14–20.
- Rusanov K, Kovacheva N, Stefanova K, et al. Rosa damascena - genetic resources and capacity building for molecular breeding. Biotechnol Biotechnol Equip. 2009;23:1436–1439.
- Staikov V, Zolotovich G. [Localization of the essential oil in the flower of Rosa damascena Miller.] Izvestia na Instituta po rastenievadstvo. 1957;IV:207–215. (in Bulgarian).
- Spiller M, Linde M, Hibrand-Saint Oyant L, et al. Towards a unified genetic map for diploid roses. Theor Appl Genet. 2011;122:489–500.
- Debener T, Mattiesch L. Construction of a genetic linkage map for roses using RAPD and AFLP markers. Theor Appl Genet. 1999;99:891–899.
- Koning-Boucoiran CF, Gitonga VW, Yan Z, et al. The mode of inheritance in tetraploid cut roses. Theor Appl Genet. 2012;125:591–607.
- Jones S. The inheritance of plant and flower traits in rose [Undergraduate Research Scholars]. College Station, TX (USA): Texas A&M University; 2013.
- Iwata H, Kato T, Ohno S. Triparental origin of Damask roses. Gene. 2000;259:53–59.
- Baydar H, Erbaş S, Kazaz S. Variations in floral characteristics and scent composition and the breeding potential in seed-derived oil-bearing roses (Rosa damascena Mill.). Turk J Agric Forestry. 2016;40:560–569.
- Rusanov K, Kovacheva N, Rusanova M, et al. Traditional Rosa damascena flower harvesting practices evaluated through GC/MS metabolite profiling of flower volatiles. Food Chem. 2011;129:1851–1859.
- Dubois A, Raymond O, Maene M, et al. Tinkering with the C-function: a molecular frame for the selection of double flowers in cultivated roses. PLoS One. 2010;5:e9288.
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3rd ed. Cold Spring Harbor, N.Y. (USA): Cold Spring Harbor Laboratory Press; 2001.
- Hibrand Saint-Oyant L, Ruttink T, Hamama L, et al. A high-quality genome sequence of Rosa chinensis to elucidate ornamental traits. Nat Plants. 2018;4:473–484.
- O’Maoileidigh DS, Graciet E, Wellmer F. Gene networks controlling Arabidopsis thaliana flower development. New Phytol. 2014;201:16–30.
- Krizek BA, Fletcher JC. Molecular mechanisms of flower development: an armchair guide. Nat Rev Genet. 2005;6:688–698.

