Abstract
Periodontal diseases, as an important part of oral pathology, have their distinctive characteristics when affecting children and young adults. The aim of this study was to identify the subgingival microorganisms through the use of real-time polymerase chain reaction (PCR) in children at the age of puberty who have plaque-induced gingivitis. The subjects of observation were 60 children, aged between 10 and 14, who did not have any systemic diseases: 30 without gingivitis (up to 25% PBI); 30 with clinically diagnosed plaque-induced gingivitis (over 25% PBI). The clinical status of each child was registered using a custom-made medical card. Gingival sulcus samples were taken with a paper pin from six teeth for real time PCR identification based on nine control strains (a comprehensive sample). Samples were transported in standardised containers. The results showed that the average quantities of the tested microorganisms were between 1 × 102 and 1 × 105 microorganisms per sample. There was a trend towards a putative increase in the average quantities of microorganisms, except for Fusobacterium nucleatum, in children with gingivitis as compared to healthy children. There were associations with up to four microorganisms in healthy children. In children with gingivitis, there was greater diversity of microorganisms, with half of the children from this group having associations with 5–7 microorganisms. The subgingival microflora became more complex in children with gingivitis: the predominant group were microorganisms from the red complex (Porphyromonas gingivalis, Treponema denticola, Tannerella forsythia) and the frequency of microorganisms from the orange complex also increased.
Introduction
Periodontal diseases, as an important part of oral pathology, are primarily chronic and have their distinctive characteristics when affecting children and young adults. Plaque-induced gingivitis is predominant during puberty. Although rarely, aggressive periodontitis can also be found in children and young adults, while chronic periodontitis can begin during puberty and progress with time. In most cases the dental biofilm is the leading etiological and/or risk factor, and the gingival sulcus is the oral niche for the development of periodontal bacterial infections [Citation1–3].
According to the current understanding of the pathogenesis of periodontal diseases, these infections are opportunistic in nature and are caused by bacteria that are part of the oral microflora. A continually high level of periodontal pathogens in the subgingival biofilm is a risk factor in developing a periodontal disease. These bacteria become pathogenic when present at a higher quantity than usual and when local immune deficiency and certain local risk factors occur [Citation4–7].
Socransky [Citation3,Citation4] is the founder of contemporary periodontal bacteriology, and the discipline has undergone rapid development due to the new methods of molecular biology [polymerase chain reaction (PCR)] for microbial identification. The subgingival microorganisms have been studied through PCR and DNA–DNA hybridisation and based on their simultaneous presence in the periodontal environment. These microorganisms have been grouped into five complexes: purple, yellow, green, orange and red [Citation3–5].
PCR identification of periodontal pathogens has ushered in a new era in periodontal science and practice [Citation8,Citation9]. There are limited data about the subgingival microorganisms in children, but it has been proved that these microorganisms undergo changes with the growth and development of the child. Subgingival microorganisms are influenced by the immune system, hormonal changes, tooth eruption, as well as by gingival inflammation. Inflammatory changes in the gingiva are significant to the formation of the subgingival biofilm and to the composition of the subgingival microflora [Citation1,Citation10].
The aim of this study was to identify the subgingival microorganisms in children with plaque-induced gingivitis, using real-time PCR. We described the periodontal status and the oral hygiene in children with and without plaque-induced gingivitis and studied their subgingival microorganisms.
Subjects and methods
Subjects
The subjects of observation were 60 children, aged between 10 and 14, who did not have any systemic diseases and had not taken any antibiotics three months prior to the study. The children were distributed into two groups: 30 children without gingivitis and with good oral hygiene (the control group) and 30 children with clinically diagnosed plaque-induced gingivitis.
Ethics statement
The study was designed and performed in compliance with the ethical principles for medical research involving human subjects in the Declaration of Helsinki. We confirm that all patients or participants (or their parent or legal guardian) gave written informed consent to the inclusion in this study and that they acknowledge that they cannot be identified via the paper and that we have fully anonymised them.
Clinical examination of the children
The periodontal status of all children was registered with the use of a medical card developed for this purpose. The following indices were taken into consideration: Oral-hygienic index of Green Vermillion (OHI-GV), Papilla Bleeding Index Saxer & Mulheman (PBI-severity).
We assumed that all children with a PBI-spread of less than 25% (percentage of bleeding papillae in all papillae studied) fall into the control group, that is, children not suffering from plaque-induced gingivitis, and the children with PBI of over 50%, into the group suffering from plaque-induced gingivitis.
Methodology for the identification of subgingival microorganisms
To identify the main subgingival microorganisms and determine their quantity (microorganisms in samples), we used real time PCR. Samples were taken from the gingival sulcus of six teeth with a paper pin: three molars, two canines and one incisor (16, 13, 11, 26, 36, 43 teeth). The samples were taken in the morning, between 9 and 10 o’clock, at least half an hour after teeth brushing and an hour after eating. Then, they were sent for testing in standardised containers. Nine control strains were tested (comprehensive sample), .
Table 1. Microorganisms tested, grouped according to Socransky.
DNA was isolated using MagNA Pure 96 from Roche Diagnostics. For DNA isolation from Gram-positive and Gram-negative bacteria, we used a kit from Roche Diagnostics. The PCR set-up was done with Hamilton’s Microlab STARlet IVD. The final analysis of the samples was done using a LightCycler II 480 (Roche).
Data analysis
Data are presented as mean values with standard deviation (±SD). Statistical analysis was performed using independent samples T-test (comparison between two groups). p < .05 indicated statistically significant differences. The results were analysed using SPSS – 19 software.
Results and discussion
Periodontal status and oral hygiene
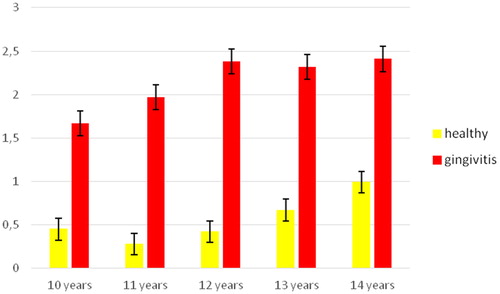
The assessment of the children’s oral hygiene using OHI-GV () showed that the quantity of plaque in children with clinically diagnosed gingivitis was significantly higher. These differences were maintained through the whole age range (t = 12.235; p < .05).
The two groups had different gingival status as evaluated on the basis of papillae bleeding (). The average PBI-severity values in the children with plaque-induced gingivitis were about fivefold higher than those in the healthy children (p > .05).
Table 2. Degree of severity of gingival inflammation.
These results indicated that the children from the test-group suffered from generalised gingivitis with a moderately severe degree of inflammation. In this group, there were significantly higher values of bleeding (PBI-severity) when compared to the healthy children from the control group.
Subgingival microorganism counts/abundance of subgingival microorganisms
The average quantities of the tested microorganisms ranged from 102 to 105 (). The average quantity of all microorganisms was significantly higher in the subjects with gingivitis as compared to the healthy children, with the exception of Fusobacterium nucleatum, whose average quantities were approximately equal in both groups of children (p > .05). The most abundant microorganism was Capnocytophaga gingivalis (green complex) – 2.5 × 105, followed by F. nucleatum – 3.7 × 104. These microorganisms participate in the processes of coaggregation, which are necessary for the development of subgingival biofilms. The quantities of Peptostreptococcus micros (1.6 × 103) were significantly lower, followed by the negligibly small quantities of Eubacterium nodatum.
Table 3. Average quantity of isolated microorganisms.
In children without gingivitis and with low quantities of plaque, there were microorganisms of the orange complex – Porphyromonas intermedia, P. micros and microorganisms important for the processes of coaggregation (F. nucleatum, C. gingivalis). Although C. gingivalis was isolated in all subjects, it was present in significantly higher quantities in the children with plaque-induced gingivitis (p < .05). P. micros, which was found in smaller quantities, was significantly higher in the children with gingivitis as compared to the healthy children (p < .05).
It is noteworthy that Porphyromonas gingivalis, which has been proved to have strong pathogenicity, was isolated in only eight of the children with gingivitis but had some of the highest average quantities – 8.2 × 104. Besides P. gingivalis, there were increased quantities (p < .05) of the other two microorganisms from the red complex (Treponema denticola, Tannerella forsythia) in the children with gingivitis as compared to the healthy children. These microbes, even in minimal quantities, are considered as a risk factor for the initiation of periodontal diseases in children in puberty. Aggreggatibacter actinomycetemcomitans was isolated only in the children with plaque-induced gingivitis, in relatively low quantities (2.9 × 103).
Our results are in agreement with previous reports. Loozen et al. [Citation10] found that the quantities of subgingival microorganisms showed a clear increase in the abundance and prevalence with increasing the clinical manifestations of inflammation. They reported that P. gingivalis, T. forsythia, T. denticola, E. nodatum, Porphyromonas micra and P. intermedia showed a clear increase in the abundance and prevalence with increasing pocket depth. Correlation matrices illustrated that almost all microorganisms were in one way correlated to other species and most of these correlations were significant.
In another study, the total amount of microorganisms detected in gingival sulcus was strongly dependent on the clinical manifestations of inflammation; it increased progressively from the healthy state to gingivitis [Citation11]. The presence of P. gingivalis can show strong association with periodontal diseases [Citation12]. However, despite the large number of A. actinomycetemcomitans-positive subjects, the subjects may not suffer from periodontal disease if the abundance of A. actinomycetemcomitans is not sufficient to cause disease [Citation13]. Consistent with the results from this study, it is generally accepted that potential pathogenic bacteria belong to the commensal oral microflora [Citation14–18].
The results from our study are similar to those recently reported by Yang et al. [Citation19], who found that P. gingivalis, P. intermedia and Tannerella forsythensis are present in greater quantities in samples taken from children with periodontal diseases, as compared to samples taken from healthy children. Yang et al. [Citation19] also found that in the group with periodontitis, F. nucleatum was in significantly greater quantities, when compared to the group of healthy children, but no significant difference in the quantities of the microorganisms was found between the group of healthy children and the group of children with gingivitis. All species of microorganisms are found in greater quantities in the groups with a more severe periodontal pathology. This suggests a possible link between the quantity of P. gingivalis, P. intermedia, T. forsythia and F. nucleatum and gingival inflammation. This is also confirmed by some authors, based on the strong correlation between the levels of the aforementioned microorganisms and the clinical indices [Citation11,Citation15–19].
F. nucleatum is one of the most studied microorganisms implicated in the aetiology of periodontal diseases. It has been isolated in the largest number of subjects, and in the highest levels [Citation13,Citation14,Citation19]. F. nucleatum is an important microorganism in the formation of dental biofilms by adhering to most plaque bacteria and contributing to the linking of early and late migratory microorganisms [Citation20]. This determines it as an intermediate coloniser and an intermediary between the beneficial microflora and the periodontally pathogenic strains [Citation19]. F. nucleatum can be isolated from individuals both with or without signs of periodontal disease [Citation13,Citation20]. However, Zhou et al. [Citation15] found that only 58.5% of healthy young adults were positive for this bacterium.
Other studies show that F. nucleatum is one of the secondary colonisers that accumulate in the biofilm. It is crucial in the interaction between the Gram-positive and Gram-negative species, and important for the bacterial colonisation that follows, through its ability to provide a link with other periodontal pathogens. Studies link this microorganism to the subgingival microflora in 27–100% of children in the prepubescent period and during a gingival inflammation the average quantities increase [Citation5,Citation10,Citation11,Citation14,Citation19].
Microbial associations
The frequency and type of the microorganisms tested in children with and without plaque-induced gingivitis were studied. We followed the number and frequency distribution of the isolated subgingival microorganisms, grouped into microbial associations, in the children ().
Table 4. Microbial associations in healthy children and children with gingivitis.
In the group of healthy children, the predominant associations were those with a lower number of microorganisms (). In the children with plaque-induced gingivitis, we observed microbial associations with a larger number of microorganisms (). In half of the healthy children tested, there were microbial associations with two types of microorganisms. In a quarter of these children, only a single microorganism was isolated. In the remaining cases, there were microbial associations of three or four microorganisms.
Figure 2. Microbial associations according to the number of microorganisms (MO) in healthy children.
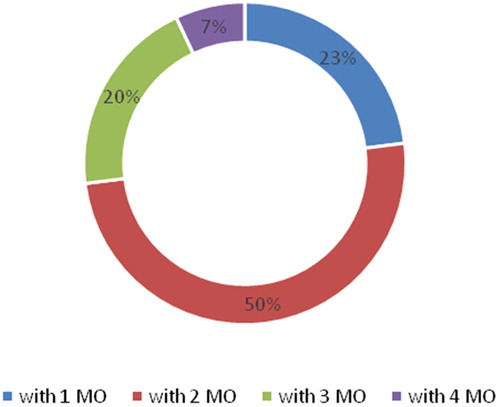
Figure 3. Microbial associations of the microorganisms (MO) in the subgingival microflora of children with plaque-induced gingivitis.
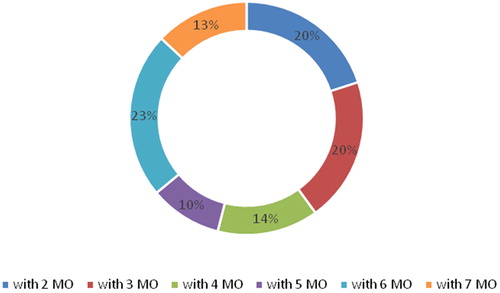
In the children with gingivitis, there was a greater diversity of microbial associations: from 2 to 7 microorganisms. Half of the children had associations with 2–4 microorganisms and the other half, with 5–7 microorganisms. The subgingival microflora became more complex when microorganisms from the red complex (P. gingivalis, T. denticola, T. forsythia) were present and the frequency of microorganisms from the orange complex increased.
We also observed a trend towards an increase in the variety of species from the red complex (T. denticola, T. forsythia and P. gingivalis). These species were seen in almost all combinations, especially in ones where there were at least three participating species. According to Ezzo and Cutler [Citation21], the presence of T. forsythia and T. denticola, even in small amounts and in isolated cases, is a risk factor for the initiation of periodontal diseases in children in puberty.
We found that A. actinomycetemcomitans was present in seven of the combinations, which also were among the more numerous microbial associations. Many authors consider that the initiation and progression of periodontal diseases are directly linked to the colonisation of microorganisms such as A. actinomycetemcomitans, as well as microorganisms from the red complex, as classified by Socransky [Citation3,Citation4]. Kulekci identified microorganism associations that are destructive to the periodontium in early childhood and in young adults [Citation20]. The present study provided evidence in support of these observations in pre-teen age children and early teens, which is a less studied age group [Citation22–26].
Conclusions
The results from this study support the conclusion that periodontal pathogens occur in childhood after the eruption of permanent teeth. Additional evidence was obtained that the quantity and variety of microobial species increases during gingival inflammation, as well as when children transition into puberty. Associations with up to four microorganisms were observed in healthy children, whereas the diversity of microorganisms was higher in children with gingivitis. Half of the children with gingivitis had associations with 5–7 microorganisms. The subgingival microflora was more complex in children with gingivitis, with microorganisms from the red complex (P. gingivalis, T. denticola, T. forsythia) being predominant and the frequency of microorganisms from the orange complex also increasing.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Rashkova M. Periodantal diseases in children and adolescents. Sofia (Bulgaria): Direct Services; 2016.
- Mascarenhas P, Gapski R, Al-Shammari K, et al. Influence of sex hormones on the periodontium. J Clin Periodontol. 2003;30:671–681.
- Socransky SS, Haffajee AD, Cugini MA, et al. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144.
- Socransky SS, Haffajee AD. Dental biofilms: difficult therapeutic targets. Periodontology 2000. 2002;28:12–55.
- Mombelli A, Gusberti EA, Oosten M. V, et al. Gingival health and gingivitis development during puberty. A 4-year longitudinal study. J Clin Periodontol. 1989;16:451–456.
- Kumar P. Sex and the subgingival microbiome: do female sex steroids affect periodontal bacteria? Periodontology 2000. 2013;61:103–124.
- Weinberg M, Westphal C, Froum S, et al. Comprehensive periodontics, 3rd ed. Boston (MA): Pearson; 2010.
- Jervøe Storm PM, Koltzscher M, Falk W. Comparison of culture and real-time PCR for detection and quantification of five putative periodontopathogenic bacteria in subgingival plaque samples. J Clin Periodontol. 2005;32:778–783.
- Marin M, Ambrosio N, Herrera D, et al. Validation of a multiplex qPCR assay for the identification and quantification of Aggregatibacter actinomycetemcomitans and Porphyromonas gingivalis: in vitro and subgingival plaque samples. Arch Oral Biol. 2018;88:47–53.
- Loozen G, Ozcelik O, Boon N, et al. Inter-bacterial correlations in subgingival biofilms: a large-scale survey. J Clin Periodontol. 2014;41:1–10.
- Scapoli L, Girardi A, Palmieri A, et al. Quantitative analysis of periodontal pathogens in periodontitis and gingivitis. J Biol Reg Homeos Ag. 2015;29:101–110.
- Rafiei M, Kiani F, Sayehmiri F, et al. Study of Porphyromonas gingivalis in periodontal diseases: a systematic review and meta-analysis. Med J Islam Repub Iran. 2017;31:62.
- Arenas Rodrigues VA, de Avila ED, Nakano V, et al. Qualitative, quantitative and genotypic evaluation of Aggregatibacter actinomycetemcomitans and Fusobacterium nucleatum isolated from individuals with different periodontal clinical conditions. Anaerobe. 2018;52:50–58.
- Papaioannou W, Gizani S, Haffajee AD, et al. The microbiota on different oral surfaces in healthy children. Oral Microbiol Immunol. 2009;24:183–189.
- Zhou X, Liu X, Li J, et al. Real-time PCR quantification of six periodontal pathogens in saliva samples from healthy young adults. Clin Oral Invest. 2015;19:937–946.
- Tanner AC, Milgrom PM, Kent RJ, et al. The microbiota of young children from tooth and tongue samples. J Dent Res. 2002;81:53–57.
- Armitage G. Comparison of the microbiological features of chronic and aggressive periodontitis. Periodontol 2000. 2010;53:70–88.
- Kolenbrander PE, Andersen RN, Blehert DS, et al. Communication among oral bacteria. Microbiol Mol Biol Rev. 2002;66:486–505.
- Yang NY, Zhang Q, Li JL, et al. Progression of periodontal inflammation in adolescents is associated with increased number of Porphyromonas gingivalis, Prevotella intermedia, Tannerella forsythensis, and Fusobacterium nucleatum. Int J Paediatr Dent. 2014;24:226–233.
- Choi H, Kim E, Kang J, et al. Real-time PCR quantification of 9 periodontal pathogens in saliva samples from periodontally healthy Korean young adults. J Periodontal Implant Sci. 2018;48:261–271.
- Ezzo PJ, Cutler CW. Microorganisms as risk indicators for periodontal disease. Periodontol 2000. 2003;32:24–35.
- Kulekci G, Leblebicioglu B, Keskin F, et al. Salivary detection of periodontopathic bacteria in periodontally healthy children. Anaerobe. 2008;14:49–54.
- Marsh PD, Moter A, Devine DA. Dental plaque biofilms: communities, conflict and control. Periodontol 2000. 2011;55:16–35.
- Holt SC, Ebersole JL. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000. 2005;38:72–122.
- Harvey JD. Periodontal microbiology. Dent Clin North Am. 2017;61:253–269.
- Drummond BK, Brosnan MG, Leichter JW. Management of periodontal health in children: pediatric dentistry and periodontology interface. Periodontol 2000. 2017;74:158–167.