Abstract
In this study, the genetic relationships of 29 grape genotypes were defined with six SSR (simple sequence repeat) markers, and 65 polymorphic bands were identified, with matrix correlation (r) of 0.79. The expected heterozygosity (He) ranged from 0.783 (VrZAG62) to 0.869 (VVMD7) and the observed heterozygosity (Ho) from 0.483 (VrZAG62) to 0.897 (VVS2). According to the results, the smaller main cluster included only one ancient cultivar, ‘Buca Razakısı’. Foça Karası’, an ancient cultivar in the large main group, was separated from other grapes varieties. The first sub-cluster (S1) formed by mainly introduced cultivars included three cultivars, ‘Cabernet Sauvignon’, ‘Cabarnet Franc’ and ‘Merlot’. The second sub-cluster (S2) was the largest group, and included 19 cultivars, ‘Semillion’, ‘Alicante Boushet’, ‘Delbele’, ‘Çeşme Pembesi’, ‘Grenache’, ‘Öküzgözü’, ‘Petit Syrah’, ‘Papaz Karası’, ‘Colombard’, ‘Harsleleh’, ‘Moiseylative’, ‘Şika’, ‘Müşküle’, ‘Ohannes’, ‘Cinsaut’, ‘Kırmızı Şam’, ‘Kozak Gemresi’, ‘Siyah Gemre’ and ‘Yuvarlak Razakı’. The third sub-cluster (S3) included four cultivars, ‘Cardinal’, ‘Italia’, ‘Hafızali’ and ‘Malbee’. ‘Hafizali is an ancient cultivar and is included in this group. The fourth sub-cluster included only one ancient cultivar, ‘Pek Üzümü’. Based on the rates of similarities of the cultivars included in this study, the highest rate was recorded for ‘Öküzgözü-Petit Syrah’ (93%) in the second sub-cluster. The results reported here are important as the first steps towards better characterization of these grape genotypes and will aid future germplasm management and breeding efforts.
Introduction
The domestication of grape (Vitis vinifera L.) is believed to have occurred around 4000 BC from a perennial wild grape originally classified as V. sylvestris in Afghanistan, Black Sea and the Caspian Sea, however a recent archaeological finding suggested 5400–5000 BC as the probable period of domestication of the grape in the mountains of Iran [Citation1,Citation2]. Turkey, which is located in two main floristic regions (Near-East and Mediterranean), has a rich genetic diversity potential for grapevines because its strategic position in the Mediterranean Sea and the heterozygotic hereditary structure of the grapevines have led to the emergence of greatly diversified varieties, types and species [Citation3–5]. According to De Andrés et al. [Citation6], the time when wild grapevines spread over Southern Europe and Western and Central Asia was during the Neolithic period. Archaeological and historical evidence indicates that grapevines were semi-domesticated in the Near East. Several studies have shown evidence supporting that secondary domestication events existed along the Mediterranean basin [Citation7]. The viticulture practices in Turkey date back to 3500 BC, providing a deep-rooted history in Anatolia [Citation8]. There are 1500 grape varieties in a germplasm collection of The National Germplasm Repository Vineyard of the Viticulture Research Institute in Turkey [Citation9].
V. vinifera originated in the Middle East and some of today’s varieties could be more than a 1000 years old, and although these varieties generated most of the remaining cultivars over the centuries, it could be impossible to trace back their origin [Citation10]. The domesticated grapevine spread over to European and North African Mediterranean countries and it is well documented that the diversity of grape varieties is very high around the world. A large number of grapevine varieties are reported in the literature. V. vinifera belongs to Family Vitaceae and genus Vitis with 60 species, including 10,000 varieties, many of which are synonyms.
The conventional methods of identification rely on ampelography based on the morphological differences between the varieties [Citation11–13], i.e. leaf shape and contours, growing shoots characteristics, flower sex, grape cluster shape and grape colour. Traditionally, morphological data have been used to characterize cultivars and to define relationships between them, but now, molecular markers and especially SSRs (simple sequence repeats) have become an essential tool for the cultivar identification of grape varieties and for clonal discrimination. Microsatellite markers are multiallelic and highly polymorphic and accurate means of detecting genetic polymorphism [Citation10,Citation14–25].
The most important indigenous grapevine varieties in Turkey are the white ‘Sultani Çekirdeksiz’ (‘Sultanina’ or ‘Thompson Seedless’, particularly for table grape production), ‘Emir’, ‘Narince’ and ‘Misket’ and the red ‘Öküzgözü’ and ‘Boğazkere’ [Citation2,Citation26–28]. Over the last two decades, many researchers have performed studies on identification, characterization and conservation of germplasm in different countries from national to regional and local levels. Preserving grapevine biodiversity, especially autochthonous grapevines is very important for the production of new varieties [Citation18,Citation29–36]. It is of particular importance to study the grapevine gene potential in Turkey and to prepare a catalogue for this potential to validate it in international contexts [Citation37]. The classification of Turkish grapevine cultivars is traditionally based on classical ampelographic characteristics according to OIV descriptors [Citation38]. However, the common morphological identification has different limitations and sometimes does not provide enough evidence for the correct identification of the accessions. Driven by the current requirements for cultivar improvement, investigating genetic relationships is of great importance for germplasm conservation, evaluation and utilization information, which are essential for future grape breeding programmes. This study was carried out to perform genetic characterization of 29 local and introduced grape genotypes in an ex situ collection of the experimental field of Horticulture Department at Agriculture Faculty, Ege University. The grape genotypes were analysed by six SSRs markers required by the ‘European Project GENRES CT96 No 08’ [Citation21], since different methods of SSR analysis may result in small deviations (1–2 bp) in allele size. In this study, the grape genotypes, including some ancient cultivars, were identified by SSR markers and their genetic relationships were analysed.
Materials and methods
Plant material
In this study, a total of 29 grapevine genotypes collected from the vineyard of the Horticulture Department at the Agriculture Faculty, in an ex situ collection of the experimental field of Ege University in Turkey, ‘38°27′32″ N and 27°13′21″ S’, were analysed. The soil was a well-drained sandy loam. The vines were approximately 12 years old and all cultivars were subjected to standard pruning, fertilization and spraying. Grape cultivars were grafted on the rootstock 41B (Millardet et de Grasset) and each vine was given a 2.5 × 3-m (vinexrow). The accession names and the basic ampelographic characteristics of grape varieties used in this study are listed in .
Table 1. Basic ampelographic characteristics of the grape cultivars used in this study.
Microsatellite analysis and grapevine microsatellite databases consulted
Six SSR markers were employed in the ‘European Project GENRES CT96 No 08’, and ‘2nd Edition of the OIV Descriptor list for grape varieties and Vitis species’ (https://www.genres.de/, http://www.oiv.int/) were used in polymerase chain reaction (PCR) studies [Citation21,Citation39]. A total of six sequence-tagged microsatellite site loci, fully characterized in earlier studies, were used as follows: VVS2 locus [Citation36], VVMD5, VVMD7, VVMD27loci [Citation37], VrZAG79, VrZAG62 loci [Citation38].
DNA extraction
DNA analysis was performed on 29 grapevine genotypes. Young leaf tissues of these genotypes were harvested in the spring. Genomic DNA was extracted using the cetyl trimethylammonium bromide (CTAB) method modified by Lodhi et al. [Citation39] and stored at −80 °C. An RNAse treatment was performed on the eluted DNA samples. The concentrations and purity of DNA were calculated using NanoDrop® ND-1000 [Citation28].
Genomic DNA was amplified by the PCR according to the following conditions: 5–10 ng of DNA template, 1× PCR buffer (Fermentas, Life Sciences), 1.5 mmol/L MgCl2 (Fermentas, Life Science), 0.2 mmol/L for each dNTP, 0.5 µmol/L forward and reverse primer, 0.25 Unit Taq DNA polymerase (Fermentas, Life Sciences, Burlington, CA) and milliQ water to 25 μL PCR final volume. Negative controls consisting of reactions without a DNA template were included. PTC-100TM (MJ Research Inc.) was used for amplification. PCR thermocycling reactions were performed with a 2-min initial denaturation/activation step, followed by 40 cycles at 92 °C for 30 s, annealing temperature (Ta = 52–56 °C) for 1 min, and 72 °C for 2 min, with a final extension step of 7 min at 72 °C [Citation28].
A DNA marker (100 bp) was used for the approximate quantification of the bands. Amplification products were observed on a 6% (w/v) polyacrylamide gel (Promega Silver Staining kit) stained using silver staining. The gel images of the bands were scanned and scored. The genetic profile of the accessions was compared to international databases in order to match each genotype to the corresponding variety when possible [Citation40].
Genetic analysis and evaluation
The number of alleles, allele frequencies, expected (He) and observed (Ho) heterozygosity and probability of identity were calculated using Cervus© 3.0.7 (Tristan Marshall 1998–2014) software [Citation41]. For cluster analysis, dendrograms for genotypes according to UPGMA (unweighted pair-group method with arithmetic average) grouping were obtained by using NTSYS-version 2.0 (Numerical Taxonomy and Multivariate Analysis System) [Citation40] statistical package program. Total gene diversity was distributed hierarchically according to differentiation observed in the UPGMA tree. Principal component analysis (PCA) was performed based on the genotypic data from the SSR markers.
Results and discussion
Genetic analysis and evaluation
Many microsatellite markers have been developed for Vitis species [Citation42]. In this study, as noted in ‘European Project GENRES CT96 No 08’, and ‘2nd Edition of the OIV Descriptor list for grape varieties and Vitis species’, six SSR loci with a minimum standard ‘core set’ – VVS2 locus [Citation36]; VVMD5, VVMD7, VVMD27 loci [Citation37]; VrZAG79, VrZAG62 loci [Citation38] – were used for the genetic analysis of the 29 grape accessions used in this study [Citation20,Citation39–41].
shows the SSR loci, the number of alleles (n), expected heterosygosity (He), observed heterosygosity (Ho), polymorphic information content (PIC) and null allele frequencies for 29 grape cultivars analysed at 6 SSR markers. The total number of alleles for the analysed six SSR loci was 65. Based on the number of alleles, the highest allele number was obtained from VVS2 (13 alleles), followed by VrZAG79 (12 alleles), VrZAG62 (11 alleles), VVMD7 (10 alleles), VVMD27 (10 alleles) loci and VVMD5 locus with 9 alleles. The results obtained in our study corroborated the findings of similar previous studies [Citation2,Citation38, Citation43–46]. The most informative loci were VVMD7 (0.869), VVS2 (0.846), with a PIC value of 0.837 and 0.814, respectively. In our research, the mean PIC values (0.7699) were lower than the values of expected heterozygosity (0.810). The expected heterozygosity of the investigated cultivars ranged from 0.783 (VrZAG62) to 0.869 (VVMD7) with a mean of 0.810, and the null allele frequencies were generally found to be close to zero or negative. The lowest observed heterozygosity (0.483) was detected at the VrZAG62 locus and the highest at the VVS2 loci (0.846). The average He value was 0.810; this value is nearly similarly the one reported by Martinez et al. [Citation47] (0.810) and Martin et al. [Citation48] (0.806) and the mean PIC value was 0.769.
Table 2. Genetic parameters for SSR loci in some local accessions and introduced grapes (V. vinifera).
Microsatellites have been widely used for cultivar identification [Citation49–51] and analysis of genetic relationships [Citation52]. Microsatellite technology has been used in grapevine genetics because they have high degree of polymorphism [Citation1].
The PIC value was calculated for each locus to evaluate the effectiveness of each marker. shows the allele sizes, heterozygotes, homozygotes, locus frequencies and null allele frequency for 29 grape cultivars analysed using SSR markers. The allele frequency was as follows: 130–187 in VVS2, 222–268 in VVMD5, 236–257 in VVMD7, 172–247 in VVMD27, 184–206 in VrZAG62 and 184–206 in VrZAG79. The most frequent alleles at each locus and their frequencies are presented in . The frequency of alleles was registered for VrZAG79 – 255 (39.66%), VVMD27 – 243 (35.48%), VVMD5 – 230 (35.48%), VrZAG62 – 195 (32.76%), VVS2 – 133 (30.31%) and VVMD7 – 238–247 (18.97%).
Table 3. SSR loci, allele, heterozygotes (Het.), homozygotes (Hom.), frequency (Freq.) and null allele frequencies for 29 grape cultivars analysed by six SSR markers.
Table 4. The most frequent alleles per locus and their frequency.
Genetic relatedness
The dendrogram shown in was constructed using UPGMA for the evaluation of the genetic diversity and relatedness between the investigated cultivars. In the dendrogram, the investigated cultivars were separated into two main clusters. The smaller main cluster included only one ancient cultivar, ‘Buca Razakısı’, which is a razaki grape variety. ‘Buca Razakısı’ is completely separated from the other grape varieties. The Turkish genetic pool was formed during thousands of years of folk selection and later was enriched by hybridizations and mutations. Local varieties are an important resource of genes for viticulture. Their genotypes may be of interest in viticulture in addition to the advantage of their adaptation to local conditions.
Figure 1. Dendrogram of hierarchical cluster analysis based on SSR markers. Note: 1: Cabernet Sauvignon; 2: Merlot; 3: Semillion; 4: Alicante Boushet; 5: Buca Razakısı; 6: Cabarnet Franc; 7: Cardinal; 8: Cinsaut; 9: Colombard; 10: Çeşme Pembesi; 11: Delbele; 12: Foça Karası; 13: Grenache; 14: Hafızali; 15: Harsleleh; 16: Italia; 17: Kırmızı Şam; 18: Kozak Gemresi; 19: Malbee; 20: Moiseylative; 21: Müşküle; 22: Ohannes; 23: Öküzgözü; 24: Papaz Karası; 25: Pek Üzümü; 26: Petit Syrah; 27: Siyah Gemre; 28: Şika; 29: Yuvarlak Razakı.
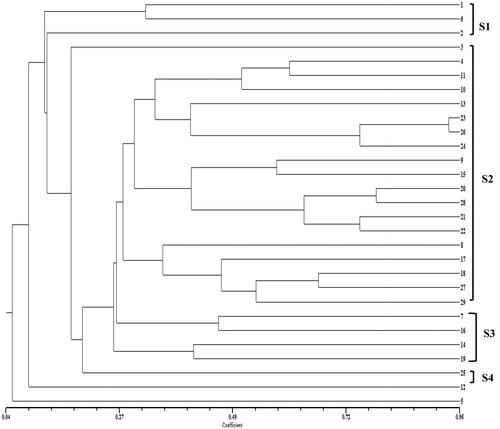
The large cluster exhibited four distinct sub-clusters labelled S1, S2, S3 and S4. ‘Foça Karası’, an ancient cultivar in this group, is separated from other grapes varieties. The first sub-cluster (S1) formed by mainly introduced cultivars and included three ones: ‘Cabernet Sauvignon’, ‘Cabarnet Franc’ and ‘Merlot’. The second sub-cluster (S2) is the largest group, including 19 cultivars: ‘Semillion’, ‘Alicante Boushet’, ‘Delbele’, ‘Çeşme Pembesi’, ‘Grenache’, ‘Öküzgözü’, ‘Petit Syrah’, ‘Papaz Karası’, ‘Colombard’, ‘Harsleleh’, ‘Moiseylative’, ‘Şika’, ‘Müşküle’, ‘Ohannes’, ‘Cinsaut’, ‘Kırmızı Şam’, ‘Kozak Gemresi’, ‘Siyah Gemre’ and ‘Yuvarlak Razakı’. The matrix correlation ‘r’ found at 0.79 and above showed reliability of the dendogram. Based on the rates of similarities of cultivars included in the study, the highest rate was recorded for ‘Öküzgözü-Petit Syrah’ (93%) in the second sub-cluster (S2). Very close relationships with high similarity were determined for ‘Öküzgözü’ and ‘Petit Syrah’ with the same pedigree or origin. ‘Öküzgözü’ (an ancient grape variety having a little flavour, a loose cluster form and a rose berry colour) and ‘Petit Syrah’ (an introduced variety having a neutral flavour, a dense cluster form and a blue-black berry colour) have different ampelographic features, but had high similarity ratios (93%), which may be associated with the gene flow that naturally occurred in ancient times. Since V. vinifera has been cultivated in the Mediterranean and Middle East areas since very old times, some varieties may be very ancient: Pinot noir, Muscat Blanc à Petits Grains, and Sultanina, for example, date back probably to one or two thousands of years ago [Citation10,Citation14,Citation25].
The third sub-cluster (S3) included four cultivars: ‘Cardinal’, ‘Italia’, ‘Hafızali’ and ‘Malbee’. ‘Hafizali’ is an ancient cultivar. The fourth sub-cluster (S4) included only one ancient cultivar, ‘Pek Üzümü’. V. vinifera growing practice spread with historic migrations of people and progression of civilization; the same flow might be assumed for cultivated varieties [Citation10]. The true origins of cultivated grape varieties can be masked because of migrations and heavy reduction of diversity [Citation10]. Special care should be taken to conserve these cultivars in Turkey for the purpose of grape breeding and production of new varieties with new potential (i.e. resistance to disease) [Citation2,Citation53]. As previously acknowledged by Bergamini et al. [Citation10], the origin assessment and pedigree reconstruction in grapevine varieties is even more difficult owing to synonymy and homonymy. In such cases, SSRs are often the markers of choice, since they can identify varieties despite changes of plant phenotype in different environments. Scientists also view SSR fingerprinting as suitable for studies into genetic relations and pedigrees, because SSR markers have co-dominant, neutral behaviour and Mendelian segregation [Citation10,Citation54,Citation55].
The PCA based on the genotypic data from the SSR markers () showed that dim-1, dim-2 and dim-3 account for 28.57%, 7.95%, and 6.36% of the overall variation, respectively. The PCA results were nearly consistent with those of the UPGMA analysis, which had very small difference between ‘Öküzgözü’ and ‘Petit Syrah’. This close genetic relationship is also shown in , and is supported by clear overlapped in the PCA (). However, there were some differences between the dendogram and the PCA plot. The PCA results separated ‘Merlot’ from group S1, which may be due to differences in the calculation methods used in the methods.
Figure 2. Biplot based on PCA for some local and introduced grapes based on SSR markers. Note: 1: Cabernet Sauvignon; 2: Merlot; 3: Semillion; 4: Alicante Boushet; 5: Buca Razakısı; 6: Cabarnet Franc; 7: Cardinal; 8: Cinsaut; 9: Colombard; 10: Çeşme Pembesi; 11: Delbele; 12: Foça Karası; 13: Grenache; 14: Hafızali; 15: Harsleleh; 16: Italia; 17: Kırmızı Şam; 18: Kozak Gemresi; 19: Malbee; 20: Moiseylative; 21: Müşküle; 22: Ohannes; 23: Öküzgözü; 24: Papaz Karası; 25: Pek Üzümü; 26: Petit Syrah; 27: Siyah Gemre; 28: Şika; 29: Yuvarlak Razakı.
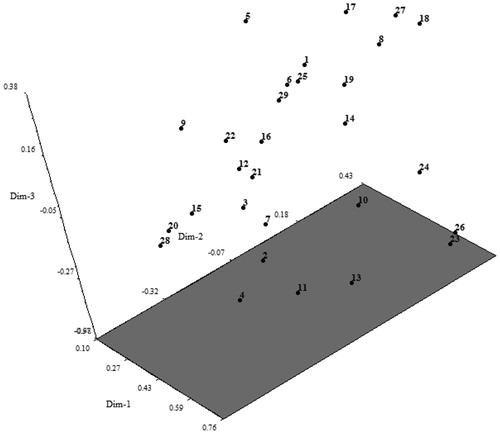
In our genetic research, mainly ancient cultivars and introduced cultivars were divided into different groups. The obtained findings uncover the intricate nature of Turkish grape cultivars, considered peculiar to the area, but possibly being the remains of ancient varieties. Therefore, the application of modern techniques like SSRs to determine the genetic relationship between the studied genotypes has been extremely important.
Conclusions
Turkey has many indigenous grapevine varieties and the genetic analysis of their cultivars is essential for the correct description of genetic resources and the ascertainment of genetic relationships between grapevine genotypes and their pedigrees. The molecular characterization and analysis of the genetic structure in the Turkish grape germplasm based on allele frequencies and heterozygosity contributes to the knowledge about the levels and the distribution of genetic diversity of the investigated cultivars. Our results confirmed the high effectiveness of using SSR-based genetic analysis of the grapevine germplasm and provide reliable information about genetic diversity and genetic relations between accessions.
Disclosure statement
No potential conflict of interest was reported by the author.
Additional information
Funding
References
- Aradhya MK, Dangl GS, Prins BH, et al. Genetic structure and differentiation in cultivated grape. Genet Res. 2003;81(3):179–192.
- Vouillamoz Jose F, McGovern PE, Ergül A, et al. Genetic characterization and relationships of traditional grape cultivars from Transcaucasia and Anatolia. Plant Genet Resour. 2006;4(2):144–158.
- Ağaoğlu YS, Çelik H. Conservation of Vitis vinifera L. germplasm in Turkey. Vignevini. 1986;13(Suppl. 12):40–42.
- Alleweldt G. Genetics of grapevine breeding. Prog Bot. 1997;58:441–454.
- Arroyo-Garcia RL, Ruiz-Garcia LB, Ocete R, et al. Genetic evidence for the existence of independent domestication events ingrapevine. Mol Ecol. 2006;15(12):3707–3714.
- De Andrés MT, Cabezas JA, Cervera MT, et al. Molecular characterization of grapevine rootstocks maintained in germplasm collections. Am J Enol Vitic. 2007;58:75–86.
- Arroyo G, Rosa A, Revilla E. The current status of wild grapevine populations (Vitis vinifera ssp sylvestris) in the Mediterranean basin. London, UK: IntechOpen Limited; 2013. Chapter 3, The Mediterranean genetic code – grapevine and olive; p. 51–72.
- Oraman MN, Ağaoğlu YS. Some characteristics of Turkey’s viticulture and the composition of its districts in viticulture. Ankara: Ankara University Agriculture Faculty Yearbook; 1969.
- Ergül A, Marasali B, Ağaoğlu YS. Molecular discrimination and identification of some Turkish grape cultivars (Vitis vinifera L.) by RAPD markers. Vitis. 2002;41(3):159–160.
- Bergamini C, Perniola R, Cardone MF, et al. The molecular characterization by SSRS reveals a new South Italian kinship and the origin of the cultivar Uva Di Troia. SpringerPlus. 2016;5(1):1562.
- Afshari S. Identification and assessment of local varieties of grapes with the method of ampelography and ampelometri, in the city of Kedar. Int Acad J Sci Eng. 2016;7(3):1–9.
- Schneider A, Marinoni Daniela T, Crespan M. Genetics and ampelography trace the origin of Muscat Fleur D'oranger. Am J Enol Vitic. 2008;59(2):200–204.
- Garcia-Muñoz S, Muñoz-Organero G, Teresa De Andrés M, et al. Ampelography – an old technique with future uses: the case of minor varieties of Vitis vinifera L. from the Balearic Islands. J Int Sci Vigne Vin. 2011;45(3):125–137.
- Bowers JE, Dangl SG, Meredith CP. Development and characterization of additional microsatellite DNA markers for grape. Am J Enol Vitic. 1999;50:243–246.
- Bessis R. Evolution of the grapevine (Vitis vinifera L.) imprinted by natural and human factors. Can J Bot. 2007;85(8):679–690.
- Regner F, Wiedeck E, Stadlbauer A. Differentiation and identification of White Riesling clones by genetic markers. Vitis. 2000;39(3):103–107.
- Riaz S, Garrison KE, Dangl GS, et al. Genetic divergence and chimerism within ancient asexually propagated winegrape cultivars. J Am Soc Hortic Sci. 2002;127(4):508–514.
- Crespan M. The parentage of Muscat of Hamburg. Vitis. 2003;42(4):193–197.
- González-Techera A, Jubany S, Ponce De León I, et al. Molecular diversity within clones of cv. Tannat (Vitis vinifera L.). Vitis. 2004;43(4):179–185.
- Hocquigny S, Pelsy F, Dumas V, et al. Diversification within grapevine cultivars goes through chimeric states. Genome. 2004;47(3):579–589.
- This P, Jung A, Boccacci P, et al. Development of a standard set of microsatellite reference alleles for identification of grape cultivars. Theor Appl Genet. 2004;109(7):1448–1458.
- Jahnke G, Májer J, Lakatos A, et al. Isoenzyme and microsatellite analysis of Vitis vinifera L. varieties from the Hungarian grape germplasm. Sci Hortic. 2009;120(2):213–221.
- Zoghlami N, Riahi L, Laucou V, et al. Origin and genetic diversity of Tunisian grapes as revealed by microsatellite markers. Sci Hortic. 2009;120(4):479–486.
- Zoghlami N, Riahi L, Laucou V, et al. Genetic structure of endangered wild grapevine Vitis vinifera ssp. sylvestris populations from Tunisia: implications for conservation and management. Forest Ecol Managet. 2013;310:896–902.
- This P, Lacombe T, Thomas MR. Historical origins and genetic diversity of wine grapes. Trends Genet.. 2006;22(9):511–519.
- Sabır A, Kafkas S, Tangolar S, et al. Genetic relationship of grape cultivars by ISSR (Inter-Simple Sequence Repeats) markers. Euro J Hort Sci. 2008;73(2):84–88.
- Sabır A, Tangolar S, Büyükalaca S, et al. Ampelographic and molecular diversity among grapevine (Vitis spp.) cultivars. Czech J Genet Plant Breed. 2009;45(4):160–168.
- Işçi B, Dilli Y. Characterization of autochthonous grapevine cultivars (Vitis vinifera L.) from the Aegean Region of Turkey using simple sequence repeats (SSRs). J Agric Sci. 2015;21:538–545.
- Gonzáles-Andrés F, Martin JP, Yuste J, et al. Identification and molecular biodiversity of autochthonous grapevine cultivars in the “Comarca del Bierzo”, Leon, Spain. Vitis. 2007;46:71–76.
- Cabezas JA, Cervera MT, Arroyo-García R, et al. Garnacha and Garnacha Tintorera: genetic relationships and the origin of teinturier varieties cultivated in Spain. Am J Enol Vitic. 2003;54(4):237–245.
- Crespan M, Calò A, Giannetto S, et al. Sangiovese and Garganega are two key varieties of the Italian grapevine assortment evolution. Vitis. 2008;47(2):97–104.
- Cunha J, Teixeira Santos M, Carneiro LC, et al. Portuguese traditional grapevine cultivars and wild vines (Vitis vinifera L.) share morphological and genetic traits. Genet Resour Crop Evol. 2009;56(7):975–989.
- De Andrès M, Benito A, Pèrez-Rivera G, et al. Genetic diversity of wild grapevine populations in Spain and their genetic relationships with cultivated grapevines. Mol Ecol. 2012;21:800–816.
- Brunori E, Cirigliano P, Biasi R. Sustainable use of genetic resources: the characterization of Italian local grapevine variety (“Grechetto Rosso”) and its own landscape. Vitis. 2015;54:261–264.
- Maletić E, Pejić I, Karoglan Kontić J, et al. Ampelographic and genetic characterization of Croatian grapevine varieties. Vitis. 2015;54:93–98.
- Thomas MR, Scott NS. Microsatellite repeats in grapevine reveal DNA polymorphisms when analysed as Sequence-Tagged Sites (STSs). Theor Appl Genet. 1993;86(8):985–990.
- Bowers JE, Dangl G, Vignani S, et al. Isolation and characterization of new polymorphic simple sequence repeat loci in grape (Vitis vinifera L.). Genome. 1996;39(4):628–633.
- Sefc KM, Regner F, Turetschek E, et al. Identification of microsatellite sequences in Vitis riparia and their applicability for genotyping of different Vitis species. Genome. 1999;42(3):367–373.
- Lodhi MA, Guang-Ning YE, Norman Weeden F, et al. A simple and efficient method for DNA extraction from grapevine cultivars and Vitis species. Plant Mol Biol Rep. 1994;2(1):6–13.
- Rohlf FJ. Phylogenetic models and reticulations. J Classif. 2000;17:185–189.
- Kalinowski ST, Taper ML, Marshall TC. Revising how the computer program cervus accommodates genotyping error increases success in paternity assignment. Mol Ecol. 2007;16(5):1099–1106.
- Samarth RR, Gaikwad MS, Anupa TPD, et al. Profilling of grape varieties using OIV descriptors varieties using OIV descriptors and molecular markers. Int Quart J Life Sci. 2016;11(4):3189–3195.
- Lopes MS, Sefc KM, Ei Ras Dias E, et al. The use of microsatellites for ge1mplasm management in a Portuguese grapevine collection. Theor Appl Genet. 1999;99(3-4):733–739.
- Fatahi R, Ebadi A, Bassil N, et al. Characterization of Iranian grapevine cultivars using microsatellite markers. Vitis. 2003;42:185–192.
- Ibañez J, Andrés MT, Molino A, et al. Genetic study of key Spanish grapevine varieties using microsatellite analysis. Am J Enol Vitic. 2003;54:22–30.
- Benjak A, Ercisli S, Vokurka A, et al. Genetic relationships among grapevine cultivars native to Crotia, Greece and Turkey. Vitis. 2005;44(2):73–78.
- Martinez LE, Cavagnaro PF, Masuelli RW, et al. SSR-based assessment of genetic diversity in South American Vitis vinifera varieties. Plant Sci. 2006;170(6):1036–1044.
- Martin JP, Borrego J, Cabello F, et al. Characterization of Spanish grapevine cultivar diversity using sequence-tagged microsatellite site markers. Genome. 2003;46(1):10–18.
- Grando MS, Frisinghelli C. Grape microsatellite markers: sizing of DNA alleles and genotype analysis of some grapevine cultivars. Vitis. 1998;37:79–82.
- Sefc K, Regner F, Glössl J, et al. Genotyping of grapevine and rootstock cultivars using microsatellite markers. Vitis. 1998;37:15–20.
- Meredith CP, Bowers JE, Riaz S, et al. The identity and parentage of the variety known in California as Petite Sirah. Am J Enol Vitic. 1999;50:236–242.
- Lefort F, Roubelakis-Angelakis K. Genetic comparison of Greek cultivars of Vitis vinifera L. by nuclear microsatellite profiling. Am J Enol Vitic. 2001;52:101–108.
- McGovern PE. Ancient wine: the search for the origins of viniculture. New Jersey, USA: Princeton University Press; 2004.
- Dakin E, Avise J. Microsatellite null alleles in parentage analysis. Heredity (Edinb). 2004;93(5):504–509.
- Atak A, Kahraman KA, Söylemezoğlu G. Ampelographic identification and comparison of some table grape (Vitis vinifera L.) clones. New Zealand J Crop Hortic Sci. 2014;42(2):77–86.
