Abstract
Genetic diversity and variation of four naturally distributed Huperzia serrata populations in Vietnam were analyzed based on the DNA fingerprint data obtained by ISSR and SCoT techniques separately and combined together. Both of ISSR and SCoT markers showed that the genetic diversity in Vietnam was relatively high at population and species level, except for the Hoang Lien population. The genetic diversity parameters of the Hoang Lien population were Nei’s gene diversity index (He) = 0.1436, Shannon index (I) = 0.2161, percentage of polymorphic bands (PPB) = 45.45%; of the Bach Ma population, He = 0.21, I = 0.33, PPB = 71.97%; of the Ngoc Linh population, He = 0.20, I = 0.31, PPB = 74.24%; of the Bidoup population, He = 0.19, I = 0.30, PPB = 74.24%; and at species level in Vietnam, He = 0.23, I = 0.36, PPB = 100%. The genetic differentiation was high with a value of GST = 0.19 and the number of migrants was estimated through gene flow as Nm = 2.16 individuals per generation among populations. The results of analysis of molecular variance (AMOVA) indicated that 28 and 72% of the total variation were among and within populations, respectively. The genetic diversity and genetic variation parameters of the investigated populations depended on the type of techniques used for DNA fingerprinting and also on the nature of each population. However, there was a high correlation and general trends among the obtained data sets when using ISSR, SCoT and the combination of these techniques.
Introduction
Huperzia serrata (Thunb. ex Murray) Trevis. (Huperziaceae) is a club moss known as a medicinal plant due to its bioactive compound, huperzine A, used for treatment of Alzheimer’s and other diseases [Citation1]. It is widely distributed in temperate and tropical zones including China, Bhutan, Cambodia, India, Indonesia, Japan, Korea, Laos, Malaysia, Myanmar, Nepal, the Philippines, Russia, Sri Lanka, Thailand, Vietnam, Australia, Central America and the Pacific islands [Citation2, Citation3]. In Vietnam, this species grows on high mountains with altitude of 1000 m or more, including Lao Cai, Cao Bang, Quang Nam, Khanh Hoa, Lam Dong provinces [Citation4]. Because of the popular belief in its healing properties, it has been overexploited in recent years. This species grows well in shadowy, wet and highly humid, rich organic matter, which restricts its habitat at higher altitudes in limited areas in Vietnam. There is evidence of continuing decline in the quality of the habitat in some locations due to the effects of habitat fragmentation, leading to a decline of H. serrata populations.
Currently, this species is considered to be endangered and its distribution is limited. There are four indigenous populations of this species distributed across four Natural conservation zones, from north to South Vietnam: Hoang Lien National Park (Lao Cai, Province), Bach Ma National Park (Thua Thien Hue Province), Ngoc Linh National Reserve (between Quang Nam and Kon Tum Province) and Bidoup National Park (Lam Dong Province), which have been identified in Vietnam. In the years 2013 and 2016, field identification research identified approximately 100–300 individual plants remaining in each of the studied regions. This record reveals that the small size of the populations and their scarce regeneration have become a major threat to the survival of the species, through the loss of genetic diversity. Such conservation and management strategies should be based on thorough knowledge of the species’ biology, especially its genetic diversity and structure within the whole distribution area. Knowledge of genetic variation and diversity within and among populations of rare and endangered species is essential for developing management strategies for both in situ and ex situ conservation activities [Citation5].
Current research methods support the use of molecular markers as suitable and accurate tools for population genetic diversity detection. To solve the above problem, the AFLP technology was used to investigate the genetic diversity within and between populations of H. serrata in China indicating a considerably high level of genetic diversity at the species level [Citation6].
However, instraspecific genetic variation studies of H. serrata have not been conducted in Vietnam. Therefore, a thorough understanding of the population genetic diversity and structure of H. serrata across range is desirable. However, studies on the genetic diversity of this species have not been conducted in Vietnam even though the reduction in genetic diversity may endanger H. serrata species here. For the long-term survival of a population or species, it is currently understood that genetic variation is critical [Citation7, Citation8]. Parameters of genetic variation and diversity within and among populations of rare and endangered species are an essential basis to develop satisfactory strategies for in situ and ex situ conservation activities [Citation5]. Thus, studies on genetic diversity and variation within and among H. serrata populations in Vietnam are necessary for biodiversity conservation and medically related uses.
The genetic diversity, variation and relationships among wild populations can be efficiently determined by using different molecular markers [Citation9]. Different molecular maker systems have their own strengths and limitations, and making the choice of maker is an important decision for studies. ISSR (Inter Simple Sequence Repeat) and SCoT (Start Codon Targeted) markers have been used successfully on a number of crops, with technical expertise, available equipment, and available research funding [Citation10, Citation11]. The ISSR technique has the advantages of low-cost, convenience of use and high level of reliability in reproducing results [Citation12–15], and has been widely used for detection of DNA polymorphism in many plant species [Citation13–18]. In addition, SCoT is another technique with properties of high reproducibility due to primer length and annealing temperature. This technique was proposed to be used in conjunction with other DNA fingerprinting techniques for applications such as genetic analysis, and quantitative trait loci mapping, especially in laboratories with a preference for agarose gel electrophoresis [Citation10]. Thus, the ISSR and SCoT markers have been successfully used for quantification of genetic diversity in different plants [Citation17–23].
The uncontrolled overharvesting of medicinal plant species such as H. serrata, Panax stipuleanatus [Citation24] and others, without an actionable conservation strategy may lead to the increased reduction of genetic diversity and reserves, much more so than indicated by the disproportion of population genetic diversities between the studied habitats. Thus, H. serrata is a critically endangered plant and it is of critical importance to further investigate this species for conservation purposes and for sustainable harvesting and use as a valuable natural resource.
In this study, the ISSR and SCoT molecular systems were employed to evaluate the genetic diversity and genetic differentiation in wild H. serrata populations found and distributed in their natural habitat. The objectives of this study were as follows: to estimate the genetic diversity; to analyze the genetic relationships and differentiation among four populations and to catalogue the data of this study for use in the conservation and sustainable utilization of the researched medicinal plants within Vietnam.
Materials and methods
Plant materials
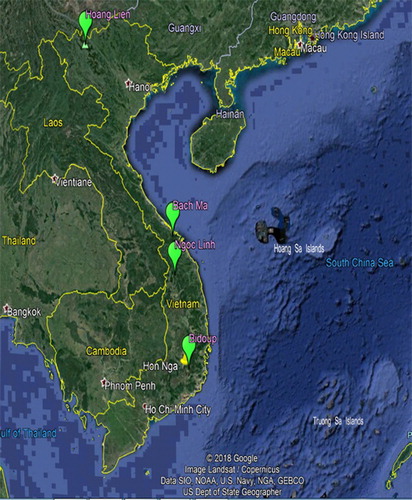
Fresh leaves were collected from 20 individuals of H. serrata per population, randomly selected from four populations distributed across four Natural conservation zones, from north to South Vietnam: Hoang Lien National Park (Lao Cai Province), Bach Ma National Park (Thua Thien Hue Province), Ngoc Linh National Reserve (between Quang Nam and Kon Tum Province) and Bidoup National Park (Lam Dong Province) (). The sampled individuals in the four populations were designated from HuL1 to HuL20 for the Hoang Lien National Park population (i.e. Hoang Lien pop.); HuB1 to HuB20 for the Bach Ma National Park population (i.e. Bach Ma pop.); HuK1 to HuK20 for Ngoc Linh National Reserve population (i.e. Ngoc Linh pop.) and HuD1 to HuD20 for Bidoup National Park population (i.e. Bidoup pop.). Fresh leaves were dried with silica gel for later DNA extraction.
DNA fingerprints
DNA fingerprints of samples were obtained by two techniques, ISSR and SCoT. ISSR primers used in this study were synthesized by PhuSa Biochem Ltd. Company (Vietnam), according to the primer set published by the University of British Columbia and Zagazig University (Egypt) [Citation25]. Twenty ISSR primers were initially screened, and 10 of them, which yielded bright, clear bands and at least possessed one polymorphic band in H. serrata populations, were chosen for DNA fingerprinting. SCoT primers used in this study were synthesized by the same company, according to the primer set published by Collard and Mackill [Citation10]. Twenty SCoT primers were initially screened, and seven of them, which yielded bright, clear bands and at least possessed one polymorphic band in H. serrata populations, were chosen for DNA fingerprinting.
DNA extraction
This study employed ISSR/SCoT makers to address genetic diversity, historical and contemporary gene flow, population bottleneck (inter population and extra population). Total genomic DNA was extracted using CTAB protocol I [Citation26] with a modification of adding 10% sodium dodecyl sulfate (SDS) to the extraction buffer which was then dissolved in water for subsequent use.
DNA fingerprints
DNA fingerprints of samples were obtained by two techniques, ISSR and SCoT. ISSR primers used in this study were synthesized by PhuSa Biochem Ltd. Company (Vietnam), according to the primer set published by the University of British Columbia and Zagazig University (Egypt) [Citation25]. Twenty ISSR primers were initially screened, and 10 of them, which yielded bright, clear bands and at least possessed one polymorphic band in H. serrata populations, were chosen for DNA fingerprinting. SCoT primers used in this study were synthesized by the same company, according to the primer set published by Collard and Mackill [Citation10]. Twenty SCoT primers were initially screened, and seven of them, which yielded bright, clear bands and at least possessed one polymorphic band in H. serrata populations, were chosen for DNA fingerprinting.
Data analysis
Since ISSR and SCoT markers are dominantly inherited, each band was assumed to represent the phenotype at a single biallelic locus [Citation27]. Microsoft Office Excel 2007 was used to estimate genetic diversity parameters: the percentage of polymorphic bands (PPB) [Citation28]. The basic parameters for genetic diversity were calculated with the POPGENE application [Citation29]. The percentage of polymorphic bands (PPB), mean Nei’s gene diversity index (He), the Shannon index (I) and the level of gene flow (Nm) [Citation30, Citation31] and Genetic distance between populations (D) were determined. Nei’s coefficient of gene differentiation (GST) [Citation32] was calculated using the Popgene 32 software: GST = (1 – HS/HT); Nm, estimate of gene flow from GST, Nm = 1/2 × (1 – GST)/GST.
The AMOVA (analysis of molecular variance) was performed through GenAlEx 6.0 program [Citation33] to describe variance components and their significance levels for variation among individuals within and among the populations. Similarity coefficient between pair of samples and UPGMA (unweighted pair group method with arithmetic mean) dendrogram for genetic relationship among studied samples was calculated and established by using NTSYSpc 2.1 (Numerical Taxonomy and Multivariate Analysis System) software [Citation34].
Results and discussion
Genetic diversity and differentiation between populations
Analysis of the DNA fingerprints induced by ISSR technique showed a total of 72 bands with an average of 7.2 bands per primer. At the species level, all of the obtained bands were polymorphic. At the population level, the mean PPB ranged from 45.83% (Hoang Lien pop.) to 81.94% (Bach Ma pop.); whereas the mean He and I ranged from 0.15 (Hoang Lien pop.) to 0.25 (Bach Ma pop.), and from 0.23 (Hoang Lien pop.) to 0.38 (Bach Ma pop.), respectively. Bach Ma population exhibited the highest PPB, He and I values, whereas Hoang Lien population did the lowest. At the species level all these parameters exhibited higher mean values, with PPB of 100%, He of 0.23 and I of 0.36 ( and ).
Table 1. Primers used in the study for DNA fingerprinting.
Table 2. Genetic diversity of H. serrata species at population and species level.
Based on the DNA fingerprints induced by SCoT technique, a total of 60 bands were recorded with an average of 8.6 bands per primer. At the species level, all of the obtained bands were polymorphic. At the population level, the mean PPB ranged from 45% (Hoang Lien pop.) to 80% (Ngoc Linh pop.); whereas the mean He and I ranged from 0.13 (Hoang Lien pop.) to 0.22 (Ngoc Linh pop.), and from 0.20 (Hoang Lien pop.) to 0.33 (Ngoc Linh pop.), respectively. Ngoc Linh population exhibited the highest PPB, He and I values, whereas Hoang Lien population, the lowest ones. At the species level, all these parameters exhibited higher mean values, with PPB of 100%, He of 0.23 and I of 0.36 ( and ).
Based on the combined data from using ISSR and SCoT techniques, PPB ranged from 45.45% (Hoang Lien pop.) to 74.24% (Ngoc Linh pop. and Bidoup pop.); whereas He ranged from 0.14 (Hoang Lien pop.) to 0.21 (Bach Ma pop.), and I from 0.22 (Hoang Lien pop.) to 0.33 (Bach Ma pop.). Ngoc Linh and Bidoup populations exhibited the highest PPB, but Bach Ma population exhibited the highest He and I value, whereas Hoang Lien population did the lowest. At the species level, all these parameters had higher mean values, with PPB of 100%, He of 0.23 and I of 0.36 ( and ).
Using the AFLP technique, Huang and He [Citation6] studied the genetic diversity of 10 H. serrata populations in China. He =0.12; I = 0.20; PPB = 53.5% were recorded for the population with the lowest levels of diversity; He = 0.20; I = 0.32; PPB = 82.0% for the population with the highest levels of diversity; and He = 0.20; I = 0.33; PPB = 86.5% for the 10 populations in total. Our findings showed a much lower level of genetic diversity in Hoang Lien population, but slightly higher level in Bach Ma, Ngoc Linh and Bidoup populations when compared to the species level in China as previously screened. As expected, our results showed that H. serrata in Vietnam exhibits relatively high level of within-population genetic variation, except the Hoang Lien population. The current forests having such populations have been greatly degraded and fragmented by the activities of local people and have formed small forest patches. All populations of H. serrata remained in such small patches; the number of observed individuals was less than 300 individuals in each population, which is consistent with the hypothesis that these areas act as small refugia for this species. They are small populations with inherently low levels of genetic variation. Furthermore, beyond the studies, the local people have been collecting plants from young to mature stages for drug purposes. This might be taken as evidence for a very low significance for vegetative propagation and clonal growth. Perhaps geitonogamy and consanguineous breeding among individuals in small populations may have reduced the within-population genetic diversity. A low level of diversity may be due to isolation resulting in loss of unexploited genetic potential. Another reason for low diversity in the marginal populations is due to the marginal position itself [Citation35]. Thus, the Hoang Lien population is of greater danger of becoming extinct than those in the South of Vietnam. Geographically, the levels of genetic diversity of H. serrata tended to increase from the North to the South of Vietnam. This may be related to geographic distribution, population size of the species, and number of tested populations. Widespread species in the South have abundant genetic variation for numerous traits likely to be involved in climatic adaption [Citation2].
In this study, the level of genetic diversity of the other three populations (Bach Ma, Ngoc Linh and Bidoup) exhibited slightly higher levels. The genetic variance within and among the investigated H. serrata populations based on ISSR, SCoT and a combination of both these techniques is shown in .
Table 3. AMOVA analysis of 80 individuals of four populations of H. serrata using ISSR, SCoT makers and a combination of these markers.
AMOVA analysis revealed a clear differentiation among all four H. serrata populations, with 26, 27 and 28% of the total genetic variability partitioned among populations and 74, 73, and 72% of the total genetic variability partitioned among individuals within populations based on ISSR, SCoT and both ISSR and SCoT in combination, respectively ( and ).
Figure 2. Genetic variation within and among investigated populations based on both ISSR and SCoT techniques.
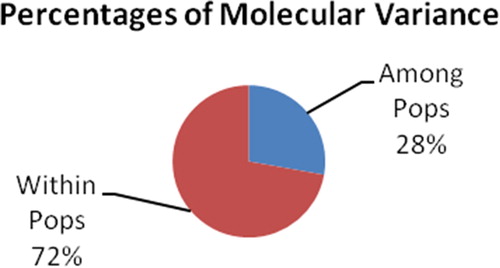
The AMOVA indicated higher genetic variation within population (72%) than among populations (28%), which suggests that H. serrata might be a heterologous plant [Citation36]. The low genetic differentiation among three of the studied populations (Bidoup, Ngoc Linh and Bach Ma) and their geographical distance suggested that H. serrata had low geographic differentiation, which could be explained on the basis of its in-crossing mode of reproduction with inbreeding. Thus, asexual reproduction results in keeping together old gene combinations, which may result in low genetic diversity [Citation19, Citation37]. Similar to observations in other species, this may be due to similar selection pressure from different locations, or insufficient interference of geographic factors towards gene follow among populations [Citation38]. Evolutionary forces such as natural selection, genetic drift, gene flow, breeding system, geographic range together with historical processes of colonization and migration are reflected in genetic variation and structure of plant populations [Citation8, Citation39].
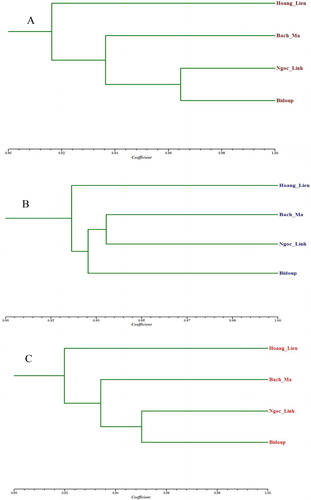
The genetic clustering pattern of H. serrata populations in Vietnam might reflect the different nature of the terrain, which would have deeply influenced the occurrence of H. serrata. Geographic and historical aspects play a role, perhaps linked to the locations of the populations in South and the population in the North on parallel foothills without direct link. When considering the genetic relationship dendrograms of the four investigated populations (), it is recognized that the trend of H. serrata species expansion was migration from the North to the South of Vietnam. The highly isolated Hoang Lien population grouped apart from the other populations. This genetic result is probably due to the decline in the habitat range. Thus, there may have been insufficient formation of next generations from the previous genetic resources in this population. The observed genetic population differentiation for H. serrata can be explained by geographic aspects and physical barriers, combined with present fragmented occurrence of populations. There is only genetic evidence of gene flow between neighboring populations. The South Vietnam accessions (Bidoup, Ngoc Linh and Bach Ma) were sourced from three regions which have a wide geographic range. These regions have a wide fluctuation in weather conditions, which may be associated with intermediate genetic diversity.
Genetic structure and relationships between four populations
Based on the DNA fingerprints obtained by the ISSR technique, the genetic differentiation among H. serrata populations was estimated to be GST = 0.18. The number of migrants was estimated as Nm = 2.24 individuals per generation among populations. Based on the SCoT technique, the genetic differentiation among populations was estimated to be GST = 0.20. The number of migrants was estimated as Nm = 2.06 individuals per generation among populations. When we combined the data obtained by these two techniques, the genetic differentiation among populations was GST = 0.19 and the number of migrants was Nm = 2.16 individuals per generation among populations.
According to the point of view of systematics of genetic differentiation, the obtained GST values showed a high genetic differentiation level [Citation27] and also high genetic differentiation comparing to the natural populations of plant taxa which possess similar characteristics to H. serrata [Citation40].
The genetic distances between the population pairs based on the combined data from two techniques in our study ranged from D = 0.05 to D = 0.09, with an average of 0.07. This range was more narrow but the average value was the same comparing with the genetic distances between the population pairs in the study of Huang and He [Citation6] (D = 0.02–0.12, with an average of 0.07). Many populations of threatened species are confided to habitat fragments with limited options for dispersal under climate change [Citation41].
These results were supported by the data for genetic distances between the pairs of investigated populations ().
Table 4. Genetic distance among investigated populations based on ISSR, SCoT markers and combined data.
The values of the genetic distances between Hoang Lien and Bach Ma populations, between Hoang Lien and Ngoc Linh populations and between Bach Ma and Bidoup populations were higher when using ISSR markers than those determined using SCoT markers. An opposite trend appeared when measuring the genetic distance between Bach Ma and Ngoc Linh populations, between Bach Ma and Bidoup populations and between Ngoc Linh and Bidoup populations. The obtained values of genetic distance between Hoang Lien and Bidoup populations were equal when using either marker. However, the general trend of genetic distances showed that Hoang Lien population was genetically closer to Bach Ma and Ngoc Linh populations than Bidoup population; Bach Ma and Ngoc Linh populations were genetically relatively close together; Bidoup and Ngoc Linh populations were genetically closest and the genetic distance between Bidoup and Bach Ma populations was medium.
The UPGMA dendrograms constructed from ISSR () and SCoT () makers and combined data () showed that the populations of H. serrata could be divided into tree geographic clusters, which corresponded to their geographic origins. The population Hoang Lien was isolated from the other ones. This population is distributed in the northern part of Vietnam. The ecotype of the habitat is the monsoon tropical climate associated with mountains, cold winter, summer rain, no dry period, and common cloudiness; the mean annual temperature is 13–16 °C with 6–8 cold months. The population Bach Ma, which is distributed in the north-central part, which has monsoon tropical climate with cool winter and summer-autumn-winter rains and common cloudiness; the mean annual temperature is 22–26 °C. The populations Ngoc Linh and Bidoup, which are distributed in Tay Nguyen plateau, grow in monsoon tropical climate associated with mountains, cold winter, autumn-summer rains, no dry period, and common cloudiness; the mean annual temperature is 15–18.5 °C with 3–4 cold months [Citation42]. It could be expected that, over the course of time, the four populations have adapted very well to the specifics of the tropical monsoon climate at these localities. The accessions of H. serrata growing in different ecological regions of Vietnam could be separated into two ecotypes depending on their geographic distribution. The Hoang Lien ecotype was clearly separated from the populations growing in the other regions. The populations from Bach Ma and Ngoc Linh, Bidoup regions belonged to a similar ecotype. These results indicated that the genetic distance of the populations of this species in Vietnam is closely associated with their geographical origin and ecological distribution.
Usability of the used techniques
Two dominant genetic markers, ISSR and SCoT, were employed to assess the levels and distribution of genetic diversity of H. serrata in Vietnam. The amplification products generated from the SCoT markers may be correlated to functional genes and their corresponding traits, while the ISSR markers detect polymorphisms in microsatellite regions that do not necessarily represent functional genes [Citation43]. Each marker system targets different regions of the genome, which leads to different sources of the detected diversity [Citation15]. Although the same number of primers were used for primer screening in ISSR and SCoT techniques, there were only 7/20 SCoT primers adapted to the criteria to choose for genetic analysis, whereas there were 10/20 ISSR primers adapted to these criteria. The number of bands amplified by the selected primers ranged from 4 to 12 with an average of 7.2 in the ISSR technique, whereas these were 7–12 with an average of 8.6 in the SCoT technique. Both of these techniques indicated low genetic diversity at population level and high genetic diversity at species level, and showed a similar clustering of H. serrata populations. The present study showed that using independent data from two different techniques can present a more efficient and accurate way to survey the genetic variability of a given species [Citation21].
Conclusions
The genetic diversity of H. serrata at population level and at species level in Vietnam was relatively high, except for population Hoang Lien Son. This genetic pattern might be due to its small population size. This information about the genetic structure provides important implications for future conservation strategies for H. serrata. All populations are of great importance for short-term and long-term survival of H. serrata in Vietnam. They should be considered as the priority subject of in situ conservation with emphasis on maintaining the number of genetically distinct individuals.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Alikhani L, Rahmani M-S, Shabanian N, et al. Genetic variability and structure of Quercus brantii assessed by ISSR, IRAP and SCoT markers. Gene. 2014;552(1):176–183.
- Anatonovis J. Genetic variation within population In: Dirzor R, Sarukan J, editors. Perspectives on plant population biology. Sunderland (MA): Sinauer; 1984. p. 229–241.
- Beardmore JA, et al. Extinction, survival and genetic variation In: Schoenwald-Cox CM, editor. Genetics and conservation. Menlo Park (CA): Benjamin-Cummings; 1983. p. 125–151.
- Bradshaw WE, Holzapfel CM. Genetic response to rapid climate change: it’s seasonal timing that matters. Mol Ecol. 2008;17(1):157–166.
- Chi VV. Vietnam medicinal plant dictionary. Ho Noi: Medicine Publish House; 1997. [Vietnamese].
- Huang J, He C. Population structure and genetic diversity of Huperzia serrata (Huperziaceae) based on amplified fragment length polymorphism (AFLP) markers. Bioch Syst Ecol. 2010;38(6):1137–1147.
- Ferrante M, Yeh J. Head and flux variability in heterogeneous unsaturated soils under transient flow conditions. Water Resour Res. 2010;5:1471–1479.
- Hogbin PM, Peakall R. Evaluation of the contribution of the genetic research to the management of the endangered plant Zieria prostrate. Conserv Biol. 1999;13(3):514–522.
- Hoffmann AA, Sgrò CM. Climate change and evolutionary adaption. Review. 2011;40:479–485.
- Collard BCY, Mackill DJ. Start codon targeted (SCoT) polymorphism: a simple, novel DNA marker technique for generating gene-targeted markers in plants. Plant Mol Biol Rep. 2009;27(1):86–93.
- Kumar P, Gupta VK, Mirsa AK, et al. Potential of molecular markers in plant biotechnology. Plant Omics J. 2009;2(4):141–162.
- Jaswinder K, Rajmeet S, et al. A systematic review on Huperzia serrata. Int J Pharmacog Phytochem Res. 2016; 8(8):1250–1255.
- Lin L, Hu Z-Y, Ni S, et al. Genetic diversity of Camellia japonica (Theaceae), a species endangered to East Asia, detected by inter-simple sequence repeat (ISSR). Biochem Syst Ecol. 2013;50:199–206.
- Lu X, Liu L, Gong Y, et al. Cultivar identification and genetic diversity analysis of broccoli and its related species with RAPD and ISSR markers. Sci Hortic. 2009;122(4):645–648.
- Luo C, He X-h, Chen H, et al. Genetic diversity of mango cultivars estimated using SCoT and ISSR markers. Biochem Syst Ecol. 2011;39(4–6):676–684.
- Ma X, Tan C, Zhu D, et al. A survey of potential huperzine A natural resources in China: the Huperziaceae. J Ethnophar. 2006;104(1–2):54–67.
- Pour-Aboughadareh A, Mahmoudi M, Moghaddam M, et al. Agro-morphological and molecular variability in Triticum boeoticum accessions from Zagros Mountains, Iran. Genet Resour Crop Evol. 2017;64(3):545–556.
- Etminan A, Pour-Aboughadareh A, et al. Evaluation of genetic diversity in a mini core collection of Iranian durum wheat germplasms. J Anim Plant Sci. 2017;27(5):1582–1587.
- Igwe DO, Afiukwa CA, Ubi BE, et al. Assessment of genetic diversity in Vigna unguiculata L. (Walp) accessions using inter-simple sequence repeat (ISSR) and start codon targeted (SCoT) polymorphic markers. BMC Genet. 2017;18(1):98–101.
- Abdel-Lateif KS, Hewedy OA. Genetic diversity among Egyptian wheat cultivars using SCoT and ISSR markers. Genet. 2018;50:36–45.
- Etminan A, Pour-Aboughadareh A, Noori A, et al. Genetic relationships and diversity among wild Salvia accessions revealed by ISSR and SCoT markers. Biotechnol Biotechnol Equip. 2018;32(3):610–617.
- Tian HZ, Han LX, Zhang JL, et al. Genetic diversity in the endangered terrestrial orchid Cypripedium japonicum in East Asia: insights into population history and implications for conservation. Sci Rep. 2018;8(1):64–67.
- Qaderi A, Omidi M, Pour-Aboughadareh A, et al. Molecular diversity and phytochemical variability in the Iranian poppy (Papaver bracteatum Lindl.): a baseline for conservation and utilization in future breeding programmes. Indust Crops Prod. 2019;130:237–247.
- Trieu LN, Mien NT, et al. Genetic diversity of Panax stipuleanatus Tsai in North Vietnam detected by Inter simple sequence repeat (ISSR) markers. Biotechnol Biotechnolog Equip. 2016;30(3):506–511.
- Admed MA. PCR techniques. Zagazig, Egypt: Department of Genetics, Zagazig University; 2005.
- McDermott JM, McDonald BA. Gene flow in plant pathosystems. Annu Rev Phytopathol. 1993;31(1):353–373.
- Nagoaka T, Ogihara Y. Applicability of inter-simple sequence repeat polymorphism in wheat for use as DNA markers in comparison to RFLP and RAPD markers. Theor Appl Genet. 1997;93:133–139.
- Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89(3):583–590.
- Nybom H, Bartish I. Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Persp Pl Ecol Evol Syst. 2000;3(2):93–114.
- Peakal R, Smouse PE. GENALEX 6: genetic analysis in Excel: population genetic software for teaching and research. Mol Ecol Notes. 2006; 6:288–195.
- Rohlf FJ. NTSYSpc numerical taxonomy and multivariate analysis system version 2.1 – user Guide. New York: Applied Biostatistics Inc.; 2004.
- Roy SC, Chakraborty BN. Genetic diversity and relationships among tea (Camellia sinensis) cultivars as revealed by RAPD and ISSR based fingerprinting. Indian J Biotechnol. 2009;8:370–376.
- Slatkin M. Gene flow in natural populations. Annu Rev Ecol S. 1985;16:393–430.
- de Vicente MC, Lopez C, et al. Genetic diversity analysis with molecular marker data: learning module. Rome, Italy: International Plant Genetic Resources Institute (IPGRI) and Cornell University; 2003.
- Hedrén M, Olofsson SN. High levels of genetic diversity in marginal populations of the marsh orchid Dactylorhiza majalis subsp. majalis. Nordic J Bot. 2018;36(4):e01747.
- Hamrick JL, Godt M. Allozyme diversity in cultivated crops. Crop Sci. 1997;37(1):26–30.
- Kumar J, Agrawal V. Assessment of genetic diversity, population structure and sex identification in dioecious crop, Trichosanthes dioica employing ISSR, SCoT and SRAP markers. Heliyon. 2019;5(3):e01346. e013462019.
- Yang J-C, Li Q-Q, Yu N, et al. Genetic diversity and structure among natural populations of Sindora glabra in Hainan Island, China as revealed by ISSR markers. Bioch Syst Ecol. 2016;69:145–151.
- Petrova G, Petrov S, Möller M, et al. Low genetic diversity in small leading edge populations of the European paleoendemic Ramonda serbica (Gesneriaceae) in Bulgaria. Nordic J Bot. 2018;36(6):njb-01655.
- Wang D-L, Qi Y-D, Feng J-D, et al. An efficient regeneration pattern via Gemmae for Huperzia serrata (Thunb. ex Murray) Trev. in Hainan Province, China. Amer Fern J. 2011;101(3):182–192.
- Zietkiewicz E, Rafalski A, Labuda D, et al. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics. 1994;20(2):176–183.
- Van NK, Hien NT, Lok PK, et al. Bioclimatic diagrams of Vietnam. Hanoi: Vietnam National University Publish House; 2000.
- Weising K, Nymbon H, Wolff K, et al. DNA fingerprinting in plants principles, methods, and applications. 2nd ed. Boca Raton (FL): CRC Press; Taylor & Francis Group; 2005.