?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
The entomopathogenic fungus Cladosporium cladosporioides is a potential candidate for biocontrol of insect pests. We isolated a strain of C. cladosporioides BOU1 from an infected brown plant hoper (BPH) of rice and characterized it using morpho-physiological and molecular analyses. Internal transcribed spacer regions and intervening 5.8S rRNA gene (ITS) sequencing and morphopathogenic analyses confirmed that BOU1 is a strain of C. cladosporioides. To select the suitable medium for this fungus, a single condium of BOU1 was grown in potato dextrose agar (PDA), potato dextrose agar with yeast (PDAY), Sabouraud dextrose agar (SDA) and synthetic nutrient-poor agar (SNA) media. The suitable medium for this fungal isolate was determined by fungal growth (colony area and conidiogenesis), and enzymatic activities (protease and lipase). The fungal growth parameters including enzymatic activities showed that the PDA medium is most suitable culture medium for C. cladosporioides. Finally, the pathogenicity of this fungal isolate was evaluated against whitefly, Bemisia tabaci through direct contact toxicity assay on eggplant leaves by dipping under laboratory conditions. The BOU1 strain caused mortality in B. tabaci in a dose-dependent manner, the highest mortality being 71% at 1 × 108 conidia/mL. To the best of our knowledge, this is the first report of isolation and molecular characterization of an entomopathogenic fungus C. cladosporioides from a BPH of rice. This study suggests that BOU1 is a potential candidate for biological control of whitefly for the promotion of sustainable agriculture.
Introduction
Cladosporium cladosporioides is a widely distributed cosmopolitan and grey pigmented mold fungus. It was isolated as a saprophyte as well as associated with plants [Citation1–3] and insects [Citation4, Citation5]. However, the identification of Cladosporium spp. only based on morphological features is difficult. Morphologically similar genera of Cladosporium spp. have already been distinguished based on morphology [Citation6–8] and DNA phylogeny [Citation9–11] of this species. Several lines of evidence suggest that C. cladosporioides is a potent entomopathogenic fungus and thus a potential candidate for biocontrol of insect pests [Citation5, Citation12].
Colony area and sporulation are important parameters in defining the virulence of entomopathogenic fungi [Citation13]. The extracellular enzymes, viz. protease and lipase, play an important role in the pathogenesis and other physiological processes of C. cladosporioides [Citation14, Citation15]; degrade the major constituents of the insect cuticle that allow hyphal penetration into the cuticle [Citation16, Citation17]. The entomopathogenic fungus C. cladosporioides has been reported to infect different species of insects such as aphid, Metopolophium dirhodum (Walker) (Hemiptera: Aphididae) [Citation18]; European pepper moth, Duponchelia fovealis (Zeller 1847) (Lepidoptera: Crambidae) [Citation19]; sweet potato whitefly, Bemisia tabaci Gennadius (Hemiptera: Aleyrodidae) [Citation4]; two-spotted spider mite, Tetranychus urticae Koch (Trombidiformes: Tetranychidae) [Citation5, Citation12]; carmine spider mite, Tetranychus cinnabarinus Boisduval (Trombidiformes: Tetranychidae) [Citation12]; and cotton bollworm, Helicoverpa armigera (Hübner 1808) [Citation20].
The present study was conducted for isolation, characterization and molecular identification of the entomopathogenic fungus C. cladosporioides from an infected brown planthopper of rice in Bangladesh. The influence of different culture media on growth and enzymatic activity of the isolated C. cladosporioides BOU1 and its pathogenicity against whitefly (B. tabaci) under laboratory conditions were also investigated. This report describes the isolation and identification of a strain of C. cladosporioides from an infected BHP of rice and discusses its potential for biocontrol of whitefly.
Materials and methods
Insect, host plant and biocontrol agent
The sweet potato whitefly, Bemisia tabaci Gennadius (Hemiptera: Aleyrodidae) biotype Asia II [Citation21], was obtained from the eggplant, Solanum melongena, field of Bangabandhu Sheikh Mujibur Rahman Agricultural University (BSMRAU), Salna, Gazipur, Bangladesh. The whitefly population was maintained on eggplant (Family - Solanaceae; variety - Baromashi Singnath Begun; seed source - Sobuj Beej Bhandar, Bangladesh). The biocontrol fungus Cladosporium cladosporioides (Davidiellaceae: Capnodiales) (Fresen.) de Vries, strain BOU1 was isolated from a surface sterilized dead brown planthopper Nilaparvata lugens Stål (Hemiptera: Delphacidae) from a farmer’s rice field in Gazipur, Bangladesh. The fungus was purified by a single conidium and cultured on potato dextrose agar (PDA) medium.
Isolation and identification of fungal isolate
Fungal isolate BOU1 was transferred into a 1.5-mL Eppendorf tube containing sterilized distilled water. Conidial suspension of 1 × 106 conidia/mL of the fungal isolate was prepared and 100-μL suspension was transferred into the Petri dishes containing 10 mL of PDA. The Petri dishes were incubated at 25 °C in a growth chamber. After three days, the dishes were checked daily until the seventh day. The fungal isolate was purified by repeated culturing of the conidia of isolated fungus growing on PDA medium. The water agar medium was also used to visualize the mycelial growth of the isolated fungus (. The morphotaxonomic characteristics of conidia-forming mycelia and conidia structure were identified [Citation22, Citation23].
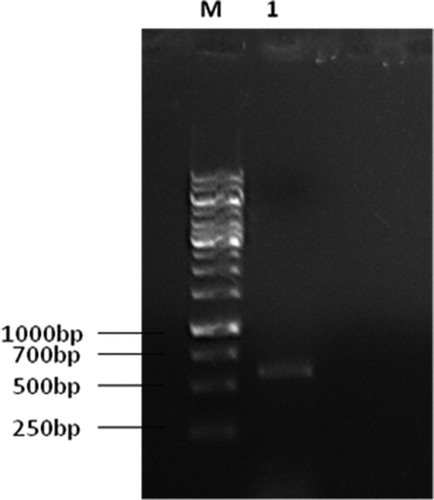
For molecular analysis, the genomic DNA of BOU1 was extracted following the modified CTAB (cetyl trimethylammonium bromide) method [Citation24]. The mycelia were harvested from the surface of plate cultures by scrapping and then ground with a mortar and pestle adding 600 µL of extraction buffer (33 mmol/L CTAB; 0.1 mol/L Tris-HCl, pH 8.0; 7.8 mmol/L ethylenediaminetetraacetic acid, EDTA; 0.7 mol/L NaCl). The mycelial suspension was collected in an Eppendorf tube and incubated at 65 °C for 30 min in a heat block with occasional shaking. The extracted DNA was precipitated with 700 µL of cold isopropanol and washed with 70% ethanol, dried under vacuum and re-suspended in TE buffer (10 mmol/L Tris-HCl, pH 8.0; 1.0 mmol/L EDTA), containing 10 µg/mL of RNase A and incubated at 37 °C for 30 min and stored at −20 °C. The nuclear rDNA region was amplified by polymerase chain reaction (PCR) [Citation25]. The PCR was performed in a 50 µL reaction mixture with DNA Taq polymerase (Promega, Madison, WI) and purified genomic DNA from the fungal isolate. The PCR amplification reaction was carried out in a thermal cycler (Veriti, Applied Biosystems, ThermoFisher Scientific) as described [Citation26]. The amplification products were subjected to electrophoresis in a 1% agarose gel and stained for 10 min in an ethidium bromide solution (10 μg/mL) and visualized under ultraviolet (UV) light (). DNA amplification was done by using the primers, ITS1 (5' TCC GTA GGT GAA CCT TGC GG 3') and ITS4 (5' TCC TCC GCT TAT TGA TAT GC 3') at the concentration of 20 pmol/µL. The amplified regions were sequenced at the National Institute of Biotechnology, Bangladesh. The nucleotide sequence of the isolate was searched for similarity index by using BLAST software. The sequence of ITS1 and ITS2 regions were subsequently submitted to GenBank.
Influence of different culture media on growth and enzymatic activity of fungal isolate
The influence of four different culture media, potato dextrose agar (PDA), potato dextrose agar with yeast (PDAY), Sabouraud dextrose agar (SDA) and synthetic nutrient-poor agar (SNA), on the growth and enzymatic activity of the BOU1 isolate of C. cladosporioides was investigated. This study was conducted by two approaches: growth (colony area and conidiogenesis) and enzymatic activities (protease and lipase) of the fungus. The measurements of colony diameter and spore production (conidiogenesis) were made daily after inoculation for 14 days. The colony area (cm2) was calculated as follows:
(1)
(1)
where, r is the radius (cm) and d is the diameter (cm) of the colony.
The activities of two enzymes, protease and lipase, of C. cladosporioides were considered as virulence of this entomopathogen. Protease activity of C. cladosporioides was determined as described by Söderhäl and Unestam [Citation27] with some modifications. One unit (U) of enzyme activity was defined as the amount of enzyme that, under the assay conditions described, gives rise to an increase of 0.1 units of absorbance in 1 h at 30 °C [Citation28]. Lipase activity of C. cladosporioides was determined as described by Pignede et al. [Citation29]. The quantity of fatty acids liberated in samples was determined by equivalents of NaOH used to reach the titration end point, accounting for any contribution from the reagent, using the following equation [Citation30]:
(2)
(2)
where, N is the normality of the NaOH titrant used (0.05 in this case). Lipase activity (U/mL) was calculated by determining the amount of supernatant that produces 1 mol of fatty acid per minute under the specified assay conditions.
Pathogenicity of whitefly by fungal isolate
The entomopathogenic effects of BOU1 were evaluated against B. tabaci through direct contact toxicity assay on eggplant leaves by dipping under laboratory conditions. When plants were 6 weeks old (4-5 true leaves), approximately 50 adult whiteflies (2 days old) were released onto each plant for 48 h to allow egg laying, after which adults were removed. The plants were then incubated for a further 12 days to allow eggs to hatch and reach the stage of second larval instars. Four suspensions of C. cladosporioides, 1 × 106, 1 × 107, 1 × 108 and 1 × 109 conidia/mL, were used against the 2nd larval instars of B. tabaci. Tween 80, 0.02% without fungal suspension was used for control. Afterwards, infested leaves were labeled and dipped into the suspensions of C. cladosporioides on both leaf surfaces for 10 s. There were 10 leaves for each treatment containing 6-10 nymphs. The numbers of living and dead nymphs were recorded (maintained at 8 days 25 ± 1 °C, 70 ± 10% RH and L 12: D 12 photoperiod) after dipping treatments on second instars were performed. Immature ones were considered dead if they had lost their normal yellow-green color, turgidity and smooth cuticle structure.
Statistical analysis
The statistical analyses were performed with a one-way analysis of variance (ANOVA) [Citation31] by Proc GLM procedure. Means were separated using Least Significant Difference (LSD) test at 5% level of significance.
Results and discussion
Based on colony morphology as well as the size and shape of conidia, the fungal isolate was initially identified as Cladosporium sp. (). The color of conidia on PDA was grey-olivaceous to dull green or olivaceous-grey, olivaceous-black, and the margins were grey-livaceous to white. Molecular analysis confirmed that BOU1 is a strain of Cladosporium cladosporioides (). The amplification of the ITS region resulted in a single product (size 507 bp) for the isolate BOU1. The GenBank accession number of the ITS1 sequence of this fungal isolate is MG654669, which was released in December 2017. The ITS1 sequence of BOU1 was very similar to those of accession numbers KY114882, KJ589542 and KJ589553 of the fungus C. cladosporioides, an average pair wise similarity of 100% (GenBank).
Figure 2. Isolated C. cladosporioides, strain BOU1: (A) infected brown planthopper (BPH) heavily colonized with BOU1; (B) growth of the infecting fungus from BPH in PDA medium at 27 °C for 7 days; (C) isolated fungal spore from the plate shown in B; (D) mycelial growth of fungus from a single spore in PDA after 24 h of incubation at 27 °C; and (E) mycelial growth of the isolated fungus at day two at 27 °C in water agar medium.
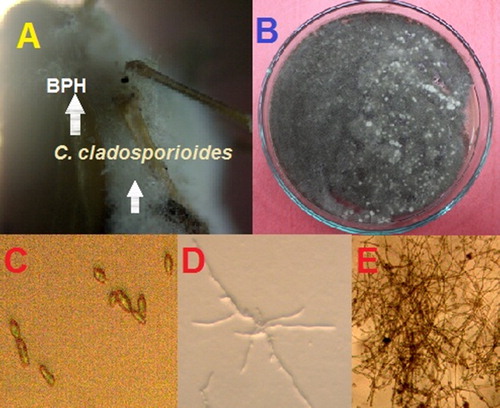
Figure 3. A phylogenetic tree constructed based on neighbor-joining method using MEGA 4.0 software. ITS1 and ITS2 regions of the seven isolates- BOU1 (This article), BFM-L47, EKF1, ZJDF16, OKF2, N37ZS and MC-24-L of Cladosporium spp. were aligned.

To find a suitable medium for culturing the fungal isolate BOU1, we tested four media, viz. PDA, PDAY, SDA and SNA, based on growth and two enzymatic activities. Significant differences were observed among the four different media on all variables, viz. colony area (F = 66.35, p < 0.0001 and df = 3 for the treatments and df = 23 for the total number of observations), conidiogenesis (F = 256.54, p < 0.0001, df = 3, 23), protease (F = 21.51, p < 0.0001 and df = 3, 23) and lipase (F = 37.49, p < 0.0001, and df = 3, 23) activities of C. cladosporioides (). The largest colony area, conidiogenesis, protease and lipase activity were 33.5 cm2, 9.9 × 105 conidia/mL, 12.8 µg/(mL h) and 9.8 µmol fatty acid/mL, respectively on PDA medium; whereas the smallest colony area, conidiogenesis, protease and lipase activity were 22.6 cm2, 0.2 × 105 conidia/mL, 10.9 µg/(mL h) and 4.1 µmol fatty acid/mL, respectively, on SNA medium (). Although the mycelium of BOU1 covered the surface area in SNA medium, the mycelium was almost invisible by the naked eye in this regard (). This indicates that the spore density of BOU1 was very low in this medium as compared with the other three media. This result suggested that SNA media is not suitable for C. cladosporioides. All the variables, viz. the colony area, conidiogenesis, protease and lipase activity, were the highest in PDA () medium; therefore, we chose PDA as the most suitable medium for in vitro cultivation of BOU1.
Figure 4. Colony area of C. cladosporioides, strain BOU1 in various media: (A) potato dextrose agar (PDA); (B) potato dextrose agar with yeast (PDAY); (C) sabouraud dextrose agar (SDA) and (D) synthetic nutrient poor agar (SNA) media.
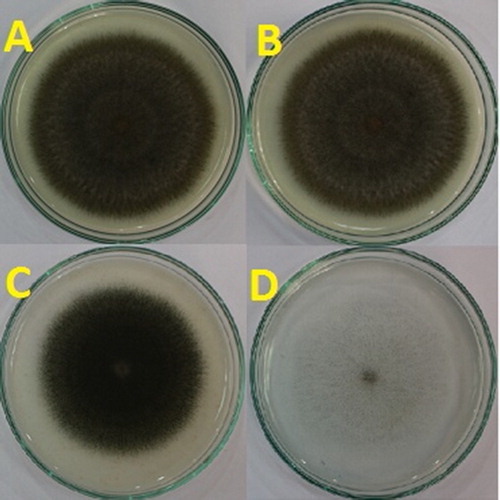
Table 1. Growth and enzyme activities of C. cladosporioides, strain BOU1 in different culture media.
The nymph mortality of B. tabaci also showed significant differences among the concentrations of C. cladosporioides (F = 1154.89, p < 0.0001 and df = 3, 23) as compared with the control (). The highest mortality (71%) of B. tabaci was recorded at 1 × 108 conidia/mL of C. cladosporioides (). The mortality percentage of B. tabaci increased with increasing the concentration of C. cladosporioides up to 1 × 108 conidia/mL; however, the mortality percentage of B. tabaci decreased when the concentration was over 1 × 108 conidia/mL (). This indicates that 1 × 108 conidia/ml concentration of BOU1 was the most effective concentration for biocontrol of whitefly.
Figure 5. Mortality rate (mean ± standard error [SE]) of whitefly after application of C. cladosporioides, strain BOU1.
Note: n, number of whitefly nymphs; same letters indicate statistically non-significant differences (LSD-test, following one-way ANOVA: P < 0.05).
![Figure 5. Mortality rate (mean ± standard error [SE]) of whitefly after application of C. cladosporioides, strain BOU1.Note: n, number of whitefly nymphs; same letters indicate statistically non-significant differences (LSD-test, following one-way ANOVA: P < 0.05).](/cms/asset/85fce333-5e3a-495e-bdaa-92f7968dd130/tbeq_a_1695541_f0005_c.jpg)
After 10 days of incubation, the culture of C. cladosporioides strain BOU1 produced a white mycelial margin with clumps of more or less verticillate branching conidiophores. These branching conidiophores became colored with the development of the spores. The conidiophores of C. Cladosporioides for a different strain are straight, solitary, unbranched, terminal or lateral and without nodules [Citation3]. These conidia are numerous, in chains of up to nine conidia and also they are limoniform, ovoid, obovoid, aseptate, light brown, hila conspicuous. The colonies of C. Cladosporioides were greenish grey to dark greenish grey, whereas the reverse of the plate revealed a greenish-black coloration [Citation32]. They also reported that the conidia of C. cladosporioides are differing in shape from ellipsoid to thin-walled limoniform, and forming long, branched, pale olive-brown chains, most without a septum, with similar coloring to conidia, and one or no septa. On its own, the morphological key used to identify species of the C. cladosporioides complex [Citation1] did not allow differentiation of the present isolates, thus it was necessary to compare this data with molecular phylogeny [Citation32]. Our results are consistent with the possible presence of cryptic species of C. cladosporioides lineages [Citation1]. A similar phenomenon was also employed for this fungus earlier [Citation3, Citation32]. The alignments and phylogenetic analysis confirmed the taxonomic identity of BOU1 with C. cladosporioides.
The germination rate of C. cladosporioides was highest on PDA medium in our investigation, in contrast to the earlier findings regarding Metarhizium anisopliae [Citation33]. Following the rate of germination, the colony area of BOU1 was also highest on PDA, which was similar to the rate of germination of M. anisopliae [Citation34]. The colony area of Isaria fumosoroseus was highest on high C/N medium [Citation33]. A similar phenomenon regarding the colony area was also reported for Beauveria bassiana and M. anisopliae [Citation35]. Our findings are also similar with the earlier findings [Citation36] that the conidiogenesis of B. bassiana was more effective on PDA medium than on other media, viz. CZA (Czapek's agar) or SDAY (Sabouraud Dextrose Agar with Yeast Extract). The BOU1 strain has high protease and lipase activities which are linked with its insect killing effects. In this study, the protease and lipase activities of C. cladosporioides were observed as a function of culture conditions and this revealed some interesting results. Both the protease and lipase activities of BOU1 were the highest on PDA medium. Previous findings indicate that the protease (Pr1) activity of M. anisopliae was highest in low and complex C/N ratio media; whereas the lipase activity of M. anisopliae was highest in 2% peptone and osmotic stress media [Citation33].
The hallmark finding of this study is that the native isolate BOU1 significantly killed a major insect pest, whitefly. The mortality rate of whitefly induced by this isolate was very high as compared to the previously reported cases in different insects, such as 64% mortality in M. dirhodum [Citation18]; 48% mortality in D. fovealis [Citation19]; and 54% mortality in H. armigera [Citation20] when using different isolates of this entomopathogen. The maximum mortality values caused by C. cladosporioides in T. urticae and T. cinnabarinus are reported to be 81.6 and 72.5% after 7 days, respectively [Citation5]. The natural infection of Cladosporium spp. on B. tabaci nymphs was 87.8% [Citation4]. This entomopathogenic fungus caused mortality of T. urticae as high as 70% within 2.77 days [Citation12]. The isolate M16 of C. cladosporioides also resulted in failure to hatch for up to 64% of H. armigera eggs [Citation20]. Our results suggest that BOU1 is a potential biocontrol agent for whitefly; therefore, it might be useful for the promotion of sustainable agriculture in Bangladesh.
Conclusions
The fungal strain BOU1 was isolated from BPH of rice in Bangladesh and initially it was characterized morpho-physiologically as Cladosporium sp. Furthermore, by using GenBank, we confirmed that the fungus is C. cladosporioides. Based on the fungal growth parameters, viz. colony area, conidiogenesis, protease and lipase activities, this research revealed that PDA is the most suitable medium for in vitro cultivation of C. cladosporioides BOU1. The BOU1 strain caused mortality in B. tabaci as high as 71% at 1 × 108 conidia/mL. Further study is needed to evaluate the biocontrol efficacy of BOU1 at both greenhouse and field levels before recommending it as a biocontrol agent for the management of whitefly (B. tabaci) in a sustainable way.
Acknowledgements
This study is a part of the postdoctoral research of Dr. Md. Touhidul Islam. He is grateful to Bangladesh Open University for providing the study leave during this research.
Funding
This study was supported by University Grant Commission (UGC) of Bangladesh as a Postdoctoral Research Fellowship to the first author and partially supported by World Bank funded sub-project no. HEQEP CP # 2071.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Bensch K, Groenewald JZ, Dijksterhuis J, et al. Species and ecological diversity within the Cladosporium cladosporioides complex (Davidiellaceae, Capnodiales). Stud Mycol. 2010;67:1–94.
- Saranya S, Ramaraju K, Jeyarani S, et al. Natural epizootics of Cladosporium cladosporioides on Tetranychus urticae Koch. (Acari.: Tetranychidae) in Coimbatore. J Biol Contr. 2013;27:95–98.
- Torres DE, Rojas-MartõÂnez RI, Zavaleta-MejõÂa E, et al. Cladosporium cladosporioides and Cladosporium pseudocladosporioides as potential new fungal antagonists of Puccinia horiana Henn., the causal agent of chrysanthemum white rust. PLoS One. 2017;12(1):e0170782. [cited 2017 Jan 31];
- Abdel-Baky NF. Cladosporium spp. An entomopathogenic fungus for controlling whiteflies and aphids in Egypt. Pakistan J Biol Sci. 2000; 3:1662–1667.
- Habashy GM, Al-Akhdar HH, Elsherbiny EA, et al. Efficacy of entomopathogenic fungi Metarhizium anisopliae and Cladosporium cladosporioides as biocontrol agents against two tetranychid mites (Acari: Tetranychidae). Egyp J Biol Pest Contr 2016;26:197–201.
- Braun U, Crous PW, Schubert K. Taxonomic revision of the genus Cladosporium s. lat. 8. Reintroduction of Graphiopsis (= Dichocladosporium) with further reassessments of cladosporioid hyphomycetes. Mycotaxon 2008; 103:207–216.
- Crous PW, Schroers HJ, Groenewald JZ, et al. Metulocladosporiella gen. nov. for the causal organism of Cladosporium speckle disease of banana. Mycol Res. 2006;110(3):264–275.
- Schubert K, Groenewald JZ, Braun U, et al. Biodiversity in the Cladosporium herbarum complex (Davidiellaceae, Capnodiales), with standardisation of methods for Cladosporium taxonomy and diagnostics. Stud Mycol. 2007; 58:105–156.
- Arzanlou A, Groenewald JZ, Gams W, et al. Phylogenetic and morphotaxonomic revision of Ramichloridium and allied genera. Stud Mycol. 2007; 58:57–93.
- de Hoog GS, Nishikaku AS, Fernandez Zeppenfeldt G, et al. Molecular analysis and pathogenicity of the Cladophialophora carrionii complex, with the description of a novel species. Stud Mycol. 2007; 58:219–234.
- Seifert KA, Hughes SJ, Boulay H, et al. Taxonomy, nomenclature and phylogeny of three cladosporium-like hyphomycetes, Sorocybe resinae, Seifertia azalea and the Hormoconis anamorph of Amorphotheca resinae. Stud Mycol. 2007; 58:235–245.
- Eken C, Hayat R. Preliminary evaluation of Cladosporium cladosporioides (Fresen.) de Vries in laboratory conditions, as a potential candidate for biocontrol of Tetranychus urticae Koch. World J Microbiol Biotechnol. 2009;25(3):489–492.
- Liu H, Skinner M, Brownbridge M, et al. Characterization of Beauveria bassiana and Metarhizium anisopliae isolates for management of tarnished plant bug, Lygus lineolaris (Himeptera, Miridae). J Invertebr Path. 2003;82(3):139–147.
- Cho EM, Boucias D, Keyhani NO. EST analysis of cDNA libraries from the entomopathogenic fungus Beauveria (Cordyceps) bassiana II. Fungal cells sporulating on chitin and producing oosporein. Microb 2006;152(9):2855–2864.
- Wang C, Hu G, St. Leger RJ. Differential gene expression by Metarhizium anisopliae growing in root exudate and host (Manducasexta) cuticle or hemolymph reveals mechanisms of physiological adaptation. Fungal Gene Biol. 2005;42(8):704–718.
- Dhawan M, Joshi N. Enzymatic comparison and mortality of Beauveria bassiana against cabbage caterpillar Pieris brassicae LINN. Brazi J Microb. 2017;48(3):522–529.
- Richard JS, Neal TD, Karl JK, et al. Model reactions for insect cuticle sclerotization: participation of amino groups in the cross-linking of Manducasexta cuticle protein MsCP36. Insect Bioche Mol Biol 2010;40:252–258.
- Abdelaziz O, Senoussi MM, Oufroukh A, et al. Pathogenicity of three entomopathogenic fungi, to the aphid species, Metopolophium dirhodum (Walker) (Hemiptera: Aphididae), and their alkaline protease activities. Egyp J Biol Pest Contr 2018;28:24. [cited 2018 Mar 08].
- Amatuzzi RF, Cardoso N, Poltronieri AS, et al. Potential of endophytic fungi as biocontrol agents of Duponchelia fovealis (Zeller) (Lepidoptera:Crambidae). Braz J Biol. 2017;78(3):429–435.
- Bahar MH, Backhouse D, PC, et al. Efficacy of a Cladosporium sp. fungus against Helicoverpa armigera (Lepidoptera: Noctuidae), other insect pests and beneficial insects of cotton. Biocontr Sci Tech. 2011;12:1387–1397.
- Khatun MSF, Jahan SMH, Lee S, et al. Genetic diversity and geographic distribution of the Bemisia tabaci species complex in Bangladesh. Acta Tropi. 2018;187:28–36.
- Humber RA. Evolution of entomopathogenicity in fungi. J Invertebr Path. 2008;98(3):262–266.
- Luangsa-Ard J, Tasanathai K, Thanakitpipattana D, et al. Novel and interesting Ophiocordyceps spp. (Ophiocordycipitaceae, Hypocreales) with superficial perithecia from Thailand. Stud Mycol. 2018;89:125–142.
- Zhang YJ, Zhang S, Liu XZ, et al. A simple method of genomic DNA extraction suitable for analysis of bulk fungal strains. Lett Appl Microb. 2010; 51:114–118.
- White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis MA, Gelfand DH, Sninsky JJ, et al., editors. PCR Protocols: A Guide to Methods and applications. San Diego, California: Academic Press; 1990. p. 315–322.
- Leong SK, Latiffah Z, Baharuddin S. Molecular characterization of Fusarium oxysporum F. Sp. cubense of banana. American J of Applied Sciences. 2009;6:1301–1307.
- Söderhäl K, Unestam T. Properties of extracellular enzymes from Amphanomyces astaci and their relevance in the penetration process of crayfish cuticle. Physiol Plant. 1975;35:140–146.
- Tremacoldi CR, Carmona EC. Production of extracellular alkaline proteases by Aspergillus clavatus. World J Microbiol Biotechnol. 2005;21(2):169–172.
- Pignede G, Wang H, Fudalej F, et al. Characterization of an extracellular lipase encoded by LIP2 in Yarrowia lipolytica. J Bacteri. 2000;182(10):2802–2810.,
- Cordenons A, Gonzalez R, Kok R, et al. Effect of nitrogen sources on the regulation of extracellular lipase production in Acinetobacter calcoaceticus strains. Biotechnol Lett. 1996;18(6):633–638.
- SAS University Edition (SAS Studio 3.6- online version). 2017. SAS Institute Inc., 100 SAS Campus Drive, Cary, NC 27513-2414, USA.
- Walker C, Muniz MFB, Rolim JM, et al. Morphological and molecular characterization of Cladosporium cladosporioides species complex causing pecan tree leaf spot. Gen Mole Res 2016;15:gmr.15038714. [cited 2016 Sep 16]. Available from: http://dx.doi.org/10.4238/gmr.15038714
- Ali S, Huang Z, Ren S. Media composition influences on growth, enzyme activity, and virulence of the entomopathogen hyphomycete Isaria fumosoroseus. Entom Exp Appl. 2009;131(1):30–38.
- Shah FA, Wang CS, Butt TM. Nutrition influences growth and virulence of the insect-pathogenic fungus Metarhizium anisopliae. FEMS Microb Lett. 2005;251(2):259–266.
- Safavi SA, Shah FA, Pakdel AK, et al. Effect of nutrition on growth and virulence of the entomopathogenic fungus Beauveria bassiana. FEMS Microb Lett. 2007;270(1):116–123.
- Ortiz-Urquiza A, Fan Y, Garrett T, et al. Growth substrates and caleosin-mediated functions affect conidial virulence in the insect pathogenic fungus Beauveria bassiana. Microb. 2016;162(11):1913–1921.