Abstract
Coronary heart disease (CHD) is a major cause of mortality and morbidity in Europe. CHD is usually caused by atherosclerosis. Despite extensive studies that have identified a large number of genetic variants, strong evidence of association with CHD are not easily replicable in different populations. Two DNA pools were constructed: one with 32 Bulgarian centenarians and one with 61 young healthy Bulgarian individuals. The pools were whole-exome sequenced and variants were annotated and quality filtered (89,810 filtered variants). Allele frequencies were estimated and Fisher’s exact test was used to evaluate the significance of allele frequency differences between the two pools. Two publicly available databases, Ensembl and DisGeNET, were used as a source of 2025 CHD-associated variants (CHD-AVs). These single nucleotide polymorphisms were screened in our data and 158 variants in 133 genes were found. ToppGene pathways analysis of genes called in both pools discovered participation of 37 genes in 9 significantly over-represented pathways. Eight variants in these 37 genes have significantly higher frequency in young individuals, and are nominated for CHD association. Variants called only in the young individuals pool (13 variants) are also nominated for association with CHD. Based on sufficiently unambiguous literature data, from the nominated variants, we prioritize five as associated with CHD in the studied Bulgarian sample. Centenarian genomes can be used to provide additional information regarding the clinical relevance of genetic variants reported as associated with CHD.
Introduction
Coronary heart disease (CHD) is responsible for approximately 20% of all deaths and almost one third of all deaths in individuals over 35 years [Citation1,Citation2]. CHD is subdivided into three distinct clinical categories: unstable angina, non-ST-segment elevation myocardial infarction and ST-segment elevation myocardial infarction. With rare exceptions, the cause of CHD is an atherosclerotic plaque (atheroma), therefore the investigation of the disease etiology relies on unraveling the molecular mechanisms of coronary atherosclerosis. Besides the proteins involved in the lipid metabolism (apolipoproteins and their receptors, PCSK9, paraoxonases, etc.), several main classes of biomolecules also play an important role in the pathogenesis of atheroma formation: scavenger receptors [Citation3–6]; growth factors and other cytokines and chemokines [Citation7–14]; adhesive molecules – integrins, cadherins, selectins and immunoglobulins [Citation15–18]; matrix metalloproteinases [Citation19–21]; nitric oxide (NO) and endothelial NO synthase [Citation22–24]; free radicals, oxidative stress and antioxidant systems [Citation23–27]; transcription factors [Citation28–31]. Atherogenesis is a complex process driven by interactions between environmental and genetic factors and their discovery and validation in individuals with different population background could impact on the prevention and risk estimation of CHD.
The aim of this study was to evaluate the carriership of genetic risk factors for predisposition to CHD in healthy Bulgarian subjects in order to prevent the development and complications of the disease.
The healthy individuals are from two different age groups, young and centenarians. The fact that centenarians have reached an extreme age indicates that their genome is devoid of pathological variants, including CHD-associated variants (CHD-AVs). For these reasons, their genomes could be considered a “gold standard” for healthy aging. It can be presumed that variants which are more frequent in the general population compared to centenarians may be associated with pathological phenotypes decreasing life expectancy. Multifactorial etiology of CHD points that known CHD-associated variants can have different effect in different populations, depending also on the environmental factors. Understanding the intra-population genetic differences between centenarians and healthy young subjects can indirectly shed light on the phenotypic effect of risk genetic variants in the given population and environmental background.
The CHD risk genetic variants in Bulgarians were determined by nomination and prioritization of CHD-AVs reported in publicly available databases and found in Bulgarian exome data. Variants were nominated for association with CHD based on their presence in genes involved in significantly over-represented pathways and statistically higher frequency among healthy individuals compared to centenarians; or based on their exclusive presence among healthy individuals. Nominated variants were then prioritized as associated with CHD based on sufficiently unambiguous literature data.
Subjects and methods
Ethics statement
The project was approved by the Ethics Committee of the Medical University of Sofia in accordance with the national and international legislation. Each DNA donor was informed about the aims of the project and signed written informed consent prior to sample collection.
Subjects
The study included a group of 32 centenarians from Bulgaria aged from 100 to 106 years (mean age 102.4) and an ethnically matched control group composed of 61 young healthy individuals from 21 to 25 years old (mean age 21.9).
Study design
Centenarians were interviewed about their lifestyle, diet, tobacco smoking, alcohol consumption, physical activity, social contacts, medical history and other important factors – positive mood, preserved memory, periods of stress, financial problems, other longevous family members.
DNA extraction and sequencing
DNA was extracted from buccal swab samples from the centenarians and blood samples from the healthy subjects. We constructed two pools: of Bulgarian centenarians and healthy young individuals with equimolar amounts of DNA from each participant. Pool sequencing was chosen as a cost-effective method for population-based studies providing reliable estimation of allele frequencies [Citation32–34]. The pools were whole-exome sequenced using BGI v4 chemistry on a BGISEQ-500 platform (by BGI Genomics) at a mean 250x coverage which is required for the detection of low frequency alleles. The obtained VCF files were annotated using the web-based ANNOVAR platform [Citation35]. The detected variants were subjected to robust filtering based on the number of individuals per pool > 30, genotype quality > 99, mapping quality > 60, number of reads per variant allele > 2, total depth of coverage > 30.
Data analysis
Ensembl genome browser [Citation36] and DisGeNet [Citation37] databases were searched for coronary heart disease associated variants. The functional significance of gene assemblages was tested using the web-based platform ToppGene [Citation38]. Pathway-based analyses allow interpretation of genes with respect to the molecular function of the product and the biological processes in which their proteins are involved [Citation39]. This approach is informative follow-up for whole exome sequencing (WES) studies, as it provides insight into the biological basis of a given phenotype. In order to minimize the false positive discoveries, the p-values were adjusted according to Benjamini and Hochberg [Citation40], and according to Benjamini and Yekutieli [Citation41].
Results and discussion
Centenarians interview data
The collected lifestyle data from the interview show in general that Bulgarian centenarians are not vegetarians (98%), most of them consume salty (84%) and sweet foods (86%), animal fats (40%). They consume fish, but not regularly, and their diet does not differ from what is typical for Bulgarians. Almost all centenarians have normosthenic habitus. It is important to note that 76% of centenarians have long lived relatives, which reflects the heritability of longevity. Another feature of almost all centenarians (93%) is their positive thinking: they are optimistic, with preserved cognitive functions and memory. Only 7% of them are smokers, but do not smoke regularly, opposed to 30% in the general population according to the National Statistical Institute (NSI) of Bulgaria. Centenarians do not consume alcohol daily or at all (63% compared to 30% in the general population). They have regular (38% vs. 9% according to NSI) or moderate physical activity (farming activity, home activity, long walks) (62% vs. 40%).
CHD-AVs found in bulgarian exome data
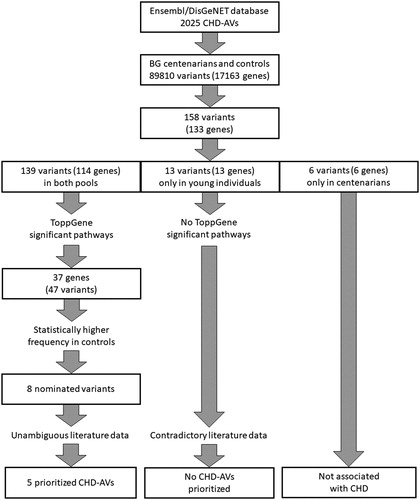
After filtering of whole-exome sequencing data, the total number of annotated variants detected in the Bulgarian pools was n = 89,810, of which in both pools n = 72,791, only in controls n = 8766 and only in centenarians n = 8253. Ensembl and DisGeNET databases list 1479 and 760 variants, respectively, as associated with CHD, yielding 2025 unique variants. Of these, 158 variants (in 133 genes) were found in Bulgarian exome data: 139 (in 114 genes) in both pools; 13 (in 13 genes) in young healthy individuals only and 6 (in 6 genes) in centenarians only.
Nomination of CHD-AVs
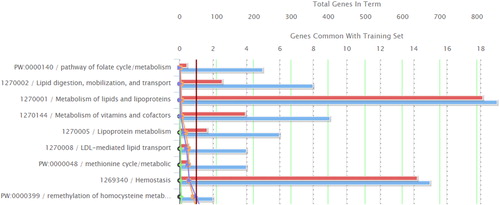
ToppGene functional enrichment analysis and candidate gene prioritization was performed using the 114 genes found in both pools. This analysis revealed 37 genes involved in nine significantly overrepresented pathways: 1. Pathway of folate cycle/metabolism; 2. Lipid digestion, mobilization, and transport; 3. Metabolism of lipids and lipoproteins; 4. Metabolism of vitamins and cofactors; 5. Lipoprotein metabolism; 6. LDL-mediated lipid transport; 7. Methionine cycle/metabolic; 8. Hemostasis; 9. Remethylation of homocysteine metabolism - cobalamin dependent (; ). These pathways could be grouped into 3 major categories: a) lipid and lipoprotein metabolism, b) hemostasis and c) vitamin and cofactor metabolism. Disturbances in the biochemical processes in these pathways could lead to the development of CHDs.
Table 1. Over-represented nine ToppGene pathways involving 37 genes out of 133 genes carrying CHD-AV in Bulgarians.
The 37 genes from the overrepresented ToppGene pathways contained 47 variants in our samples (). Eight of these variants in 7 genes (rs5443 in GNB3, rs1129293 in PIK3CG, rs693 in APOB, rs20455 in KIF6, rs1801131 and rs1801133 in MTHFR, rs174546 in FADS1, and rs2000813 in LIPG) showed statistically higher frequencies in the control pool compared to the centenarian pool.
Table 2. Allele frequencies of 47 CHD-AVs in 37 genes from over-represented ToppGene pathways in both Bulgarian pools.
The thirteen variants found only in young individuals can be speculated to have a possible negative impact on longevity. Toppgene analysis of their genes found no significantly overrepresented pathways.
The workflow of CHD-AV nomination and prioritization is summarized in .
Prioritization of variants called in both pools
Based on the significantly higher allele frequency in the young individuals, 8 variants in the genes from these pathways were nominated for CHD association: rs5443, rs1129293, rs693, rs20455, rs1801131, rs174546, rs1801133 and rs2000813. After a literature review, we prioritize 5 variants that have well established association with the pathogenesis of CHD.
The prioritization of rs5443 in the GNB3 gene, encoding G protein beta-3 subunit, is supported by literature data for its influence in the lipid metabolism and its role as an independent risk factor for hard coronary events [Citation42,Citation43].
Another nominated variant is rs693 in the APOB (apolipoprotein B) gene, which is one of the most commonly studied genes for familial hypercholesterolemia. This variant shows significant association with increased levels of circulating atherogenic lipoproteins, thus conferring increased risk for CHD [Citation44,Citation45].
Among the nominated variants there are two in the MTHFR (methylenetetrahydrofolate reductase) gene: rs1801131 and rs1801133. The MTHFR enzyme is a major participant in the folate metabolism that interlinks with various processes of DNA synthesis, repair and methylation [Citation46]. These variants are frequently found in linkage; they both lead to reduced enzyme activity of the product. The rs1801133 variant seems to have significantly greater influence, but the linkage hinders the discrimination of their individual effects [Citation47]. The variant rs1801133 has been identified as a risk factor for CHD, possibly and partly through dyslipidemic mechanisms [Citation48]. Taking the above stated arguments into account, we prioritize both rs1801131 and rs1801133 for association with CHD.
The rs174546 variant is a prominent single nucleotide polymorphism (SNP) in the FADS1 gene encoding fatty acid desaturase 1, which catalyzes a rate-limiting step in the metabolism of dietary n-3 and n-6 polyunsaturated fatty acids (PUFA) [Citation49]. The variant is associated with altered FADS1 expression leading to significant changes in the serum fatty acid ratios and increased plasma triacylglycerol levels [Citation50,Citation51]. Based on the literature data, we accept this variant as associated with CHD.
Despite the significantly higher allele frequency in the young individuals, the remaining 3 nominated variants (rs1129293, rs20455 and rs2000813) were not prioritized for CHD predisposition. The rs1129293 in the PIK3CG (phosphoinositide-3-kinase, catalytic, gamma) gene is reported to be associated with poor responsiveness to clopidogrel and decreased plasma HDL-cholesterol without metabolic or inflammatory modifications [Citation52]. It thus seems to be more relevant to drug response than to the risk of CHD. The rs20455 variant in KIF6 (Kinesin Family Member 6) is a potential pharmacogenetic variant, useful for predicting the response to statin therapy in individuals with dyslipidemia [Citation53]. Though some studies have identified an association with CHD, it does not seem to be associated with the pathogenesis per se, as pooled and meta-analyses fail to replicate the positive finding [Citation54,Citation55]. The rs2000813 in LIPG (lipase G) gene is associated with normal/moderately decreased enzyme activity leading to normal/increased HDL-C levels which are associated with protective effect against atherosclerotic cardiovascular disease [Citation56–58].
Prioritization of variants called in the young healthy individuals Pool only
None of the 13 variants from Ensemble/DisGeNET called in the young healthy individuals pool only was prioritized to be associated with CHD in Bulgarians.
Literature review of these 13 variants gives contradictory results about their functional significance. One study has found significant association of the variant rs2230806 in the ABCA1 gene with susceptibility to CHD [Citation59], whereas other studies establish this variant to have a protective role against CHD risk [Citation60,Citation61]. There are no supportive literature data of the role of rs2271570 in the C4orf33 gene as a CHD risk factor. The ROS1 gene is a proto-oncogene receptor tyrosine kinase, and the T allele of rs529038 was not found in centenarians, probably due to its oncogenic-related negative effect on longevity. Data from a meta-analysis did not identify an association of rs1801157 in the CXCL12 gene with CHD [Citation62]. For the rs10509681 variant in the CYP2C8 gene, there is no sufficient data for association with CHD. BDNF encodes a protein that plays a role in the regulation of synaptic plasticity and neuronal connections, and the variant rs6265 is reported as benign. There are insufficient studies on the association of rs14259 in PSMD9, rs1805015 in IL4R, rs255049 in DPEP3, rs2015086 in CCL18, rs2074158 in DHX58, rs1135889 in FBF1 and rs132875 in NUP50 with predisposition to CHD.
Variants called in the centenarian Pool only
The six variants found only in centenarians are as follows: rs5065 in NPPA, rs1058793 in ATP10D, rs9449444 in IBTK, rs2066714 in ABCA1, rs11838267 in C1S, and rs11881940 in HNRNPUL1. They are not expected to reduce life expectancy. Two are classified as benign in ClinVar, whereas the remaining 4 are not reported at all. None of these 6 variants could be nominated for association with CHD in the Bulgarian population.
Conclusions
CHD is determined by complex interactions between genetic and environmental factors, both of which vary across populations. The clinical relevance of reported CHD-AVs is complicated by population specific differences in their frequency and phenotype associations. Out of 2025 CHD-AVs from Ensemble/DisGeNet, only 158 were found in Bulgarian exomes of healthy individuals. Only 5 of these were prioritized for association with CHD after applying the following criteria: participation of these variants’ genes in significantly overrepresented pathways, significantly higher allele frequency in the young individuals compared to the centenarian pool, and sufficient support of our results from other studies.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Sanchis-Gomar F, Perez-Quilis C, Leischik R, et al. Epidemiology of coronary heart disease and acute coronary syndrome. Ann Transl Med. 2016;4(13):256.
- Ferreira-González I. The epidemiology of coronary heart disease. Rev Esp Cardiol. 2014;67(2):139–144.
- Kzhyshkowska J, Neyen C, Gordon S. Role of macrophage scavenger receptors in atherosclerosis. Immunobiology 2012;217(5):492–502.
- Linton MF, Tao H, Linton EF, et al. SR-BI: a multifunctional receptor in cholesterol homeostasis and atherosclerosis. Trends Endocrinol Metab. 2017;28(6):461–472.
- Park YM. CD36, a scavenger receptor implicated in atherosclerosis. Exp Mol Med. 2014;46:e99.
- Pothineni NVK, Karathanasis SK, Ding Z, et al. LOX-1 in atherosclerosis and myocardial ischemia: biology, genetics, and modulation. J Am Coll Cardiol. 2017;69(22):2759–2768.
- Fatkhullina AR, Peshkova IO, Koltsova EK. The role of cytokines in the development of atherosclerosis. Biochemistry Moscow. 2016;81(11):1358–1370.
- Hartman J, Frishman WH. Inflammation and atherosclerosis: a review of the role of interleukin-6 in the development of atherosclerosis and the potential for targeted drug therapy. Cardiol Rev. 2014;22(3):147–151.
- Hu W, Huang Y. Targeting the platelet-derived growth factor signalling in cardiovascular disease. Clin Exp Pharmacol Physiol. 2015;42(12):1221–1224.
- Libby P. Interleukin-1 beta as a target for atherosclerosis therapy: biological basis of CANTOS and beyond. J Am Coll Cardiol. 2017;70(18):2278–2289.
- Oikonomou E, Economou EK, Tousoulis D, et al. Inflammatory cytokines in atherosclerosis: current therapeutic approaches. Eur Heart J. 2016;37(22):1723–1732.
- van der Vorst EPC, Döring Y, Weber C. Chemokines and their receptors in atherosclerosis. J Mol Med. 2015;93(9):963–971.
- Xie W, Li L, Zheng X-L, et al. The role of Krüppel-like factor 14 in the pathogenesis of atherosclerosis. Atherosclerosis 2017;263:352–360.
- Zeng L, Dang TA, Schunkert H. Genetics links between transforming growth factor β pathway and coronary disease. Atherosclerosis 2016;253:237–246.
- Chistiakov DA, Orekhov AN, Bobryshev YV. Endothelial PECAM-1 and its function in vascular physiology and atherogenic pathology. Exp Mol Pathol. 2016;100(3):409–415.
- Finney AC, Stokes KY, Pattillo CB, et al. Integrin signaling in atherosclerosis. Cell Mol Life Sci. 2017;74(12):2263–2282.
- McEver RP. Selectins: initiators of leucocyte adhesion and signalling at the vascular wall. Cardiovasc Res. 2015;107(3):331–339.
- Schmidt EP, Kuebler WM, Lee WL, et al. Adhesion molecules: master controllers of the circulatory system. Compr Physiol. 2016;6(2):945–973.
- Brown BA, Williams H, George SJ. Evidence for the involvement of matrix-degrading metalloproteinases (MMPs) in atherosclerosis. Prog Mol Biol Transl Sci. 2017;147:197–237.
- Ruddy JM, Ikonomidis JS, Jones JA. Multidimensional contribution of matrix metalloproteinases to atherosclerotic plaque vulnerability: multiple mechanisms of inhibition to promote stability. J Vasc Res. 2016;53(1-2):1–16.
- Vacek TP, Rehman S, Neamtu D, et al. Matrix metalloproteinases in atherosclerosis: role of nitric oxide, hydrogen sulfide, homocysteine, and polymorphisms. Vasc Health Risk Manag. 2015;11:173–183.
- Dimitris T, Anna-Maria K, Costas Tentolouris Nikolaos P, et al. The role of nitric oxide on endothelial function. Curr Vasc Pharmacol. 2012;10(1):4–18.
- Förstermann U, Xia N, Li H. Roles of vascular oxidative stress and nitric oxide in the pathogenesis of atherosclerosis. Circ Res. 2017;120(4):713–735.
- Li H, Horke S, Förstermann U. Vascular oxidative stress, nitric oxide and atherosclerosis. Atherosclerosis 2014;237(1):208–219.
- El Assar M, Angulo J, Rodríguez-Mañas L. Oxidative stress and vascular inflammation in aging. Free Radic Biol Med. 2013;65:380–401.
- Fukai T, Ushio-Fukai M. Superoxide dismutases: role in redox signaling, vascular function, and diseases. Antioxid Redox Signal. 2011;15(6):1583–1606.
- Kattoor AJ, Pothineni NVK, Palagiri D, et al. Oxidative stress in atherosclerosis. Curr Atheroscler Rep. 2017;19(11):42.
- Chinetti-Gbaguidi G, Staels B. PPARβ in macrophages and atherosclerosis. Biochimie 2017;136:59–64.
- Jakobs P, Serbulea V, Leitinger N, et al. Nuclear factor (erythroid-derived 2)-like 2 and thioredoxin-1 in atherosclerosis and ischemia/reperfusion injury in the heart. Antioxid Redox Signal. 2017;26(12):630–644.
- Yu X-H, Zheng X-L, Tang C-K. Chapter 7: Peroxisome proliferator-activated receptor α in lipid metabolism and atherosclerosis. In: Makowski GS, editor. Advances in clinical chemistry. Vol. 71. Amsterdam: Elsevier; 2015. p. 171–203.
- Yu X-H, Zheng X-L, Tang C-K. Chapter 1: Nuclear factor-κB activation as a pathological mechanism of lipid metabolism and atherosclerosis. In: Makowski GS, editor. Advances in clinical chemistry. Vol. 70: Amsterdam: Elsevier; 2015. p. 1–30.
- Kirov G, Zaharieva I, Georgieva L, et al. A genome-wide association study in 574 schizophrenia trios using DNA pooling. Mol Psychiatry. 2009;14(8):796–803.
- Popp B, Ekici AB, Thiel CT, et al. Exome pool-seq in neurodevelopmental disorders. Eur J Hum Genet. 2017;25(12):1364–1376.
- Schlötterer C, Tobler R, Kofler R, et al. Sequencing pools of individuals — mining genome-wide polymorphism data without big funding. Nat Rev Genet. 2014;15(11):749–763.
- Yang H, Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc. 2015;10(10):1556–1566.
- Zerbino DR, Achuthan P, Akanni W, et al. Ensembl 2018. Nucleic Acids Res. 2018;46(D1):D754–D761.
- Piñero J, Bravo À, Queralt-Rosinach N, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45(D1):D833–D9.
- Chen J, Bardes EE, Aronow BJ, et al. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37(Web Server issue):W305–W11.
- Pita-Juárez Y, Altschuler G, Kariotis S, et al. The pathway coexpression network: revealing pathway relationships. PLoS Comput Biol. 2018;14(3):e1006042.
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289–300.
- Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Statist. 2001;29(4):1165–1188.
- Frey UH, Moebus S, Mohlenkamp S, et al. GNB3 gene 825 TT variant predicts hard coronary events in the population-based Heinz Nixdorf Recall study. Atherosclerosis 2014;237(2):437–442.
- Hayakawa T, Takamura T, Abe T, et al. Association of the C825T polymorphism of the G-protein beta3 subunit gene with hypertension, obesity, hyperlipidemia, insulin resistance, diabetes, diabetic complications, and diabetic therapies among Japanese. Metabolism 2007;56(1):44–48.
- Niu C, Luo Z, Yu L, et al. Associations of the APOB rs693 and rs17240441 polymorphisms with plasma APOB and lipid levels: a meta-analysis. Lipids Health Dis. 2017;16(1):166.
- Karami F, Salahshourifar I, Houshmand M. The study of rs693 and rs515135 in APOB in people with familial hypercholestrolemia. Cell J. 2019;21(1):86–91.
- Shan J-G, Xue S. MTHFR C677T polymorphism and coronary artery disease risk in the Chinese population: a meta-analysis based on 33 studies. Int J Clin Exp Med 2016;9(2):2822–2830.
- Ericson U, Sonestedt E, Ivarsson MI, et al. Folate intake, methylenetetrahydrofolate reductase polymorphisms, and breast cancer risk in women from the Malmo Diet and Cancer cohort. Cancer Epidemiol Biomarkers Prev. 2009;18(4):1101–1110.
- Luo Z, Lu Z, Muhammad I, et al. Associations of the MTHFR rs1801133 polymorphism with coronary artery disease and lipid levels: a systematic review and updated meta-analysis. Lipids Health Dis. 2018;17(1):191.
- Schuchardt JP, Köbe T, Witte V, et al. Genetic variants of the FADS gene cluster are associated with erythrocyte membrane LC PUFA levels in patients with mild cognitive impairment. J Nutr Health Aging. 2016;20(6):611–620.
- Hallmann J, Kolossa S, Gedrich K, et al. Predicting fatty acid profiles in blood based on food intake and the FADS1 rs174546 SNP. Mol Nutr Food Res. 2015;59(12):2565–2573.
- Bokor S, Dumont J, Spinneker A, et al. Single nucleotide polymorphisms in the FADS gene cluster are associated with delta-5 and delta-6 desaturase activities estimated by serum fatty acid ratios. J Lipid Res. 2010;51(8):2325–2333.
- Kächele M, Hennige AM, Machann J, et al. Variation in the phosphoinositide 3-kinase gamma gene affects plasma HDL-cholesterol without modification of metabolic or inflammatory markers. PLoS One. 2015;10(12):e0144494.
- Hamidizadeh L, Haji Hosseini Baghdad Abadi R, Babaee Baigi MA, et al. Impact of KIF6 polymorphism rs20455 on coronary heart disease risk and effectiveness of statin therapy in 100 patients from Southern Iran. Arch Iran Med. 2015;18(10):683–687.
- Li Y, Chen Z, Song H. Association between KIF6 rs20455 polymorphism and the risk of coronary heart disease (CHD): a pooled analysis of 50 individual studies including 40,059 cases and 64,032 controls. Lipids Health Dis. 2018;17(1):4.
- Vishnuprabu D, Geetha S, Bhaskar L, et al. Genotyping and meta-analysis of KIF6 Trp719Arg polymorphism in South Indian Coronary Artery Disease patients: A case-control study. Meta Gene. 2015;5:129–134.
- Vergeer M, Cohn DM, Boekholdt SM, et al. Lack of association between common genetic variation in endothelial lipase (LIPG) and the risk for CAD and DVT. Atherosclerosis 2010;211(2):558–564.
- Badellino KO, Wolfe ML, Reilly MP, et al. Endothelial lipase concentrations are increased in metabolic syndrome and associated with coronary atherosclerosis. PLoS Med. 2005;3(2):e22.
- Edmondson AC, Brown RJ, Kathiresan S, et al. Loss-of-function variants in endothelial lipase are a cause of elevated HDL cholesterol in humans. J Clin Invest. 2009;119(4):1042–1050.
- Wang F, Ji Y, Chen X, et al. ABCA1 variants rs2230806 (R219K), rs4149313 (M8831I), and rs9282541 (R230C) are associated with susceptibility to coronary heart disease. J Clin Lab Anal. 2019;33(6):20.
- Ma XY, Liu JP, Song ZY. Associations of the ATP-binding cassette transporter A1 R219K polymorphism with HDL-C level and coronary artery disease risk: a meta-analysis. Atherosclerosis 2011;215(2):428–434.
- Ghaznavi H, Aali E, Soltanpour MS. Association study of the ATP - binding cassette transporter A1 (ABCA1) Rs2230806 genetic variation with lipid profile and coronary artery disease risk in an Iranian population. Open Access Maced J Med Sci. 2018;6(2):274–279.
- Wu N, Zhang X, Jia P, et al. Lack of an association between the SDF-1 rs1801157 polymorphism and coronary heart disease: a meta-analysis. Sci Rep. 2015;5(1):11803.