Abstract
The objective of this study was isolation of humanized single-chain variable fragment (scFv) antibodies against IL1RAP, a promising cancer marker in leukemia and other tumors. Here, we screened for antibody fragments that recognized IL1RAP using a phage-display library of humanized scFv through several rounds of bio-panning and further FACS selection of enriched yeast-display sub-library. Two specific scFv clones 02 and 61 were isolated with high affinity against IL1RAP. Selected IL1RAP scFv antibodies containing Fc segment were expressed and purified from 293F cells. scFv-02 expressed and purified from mammalian cells exhibited higher IL1RAP binding affinity on IL1RAP-positive 293F cell surface compared to scFv-61. In addition, scFv-02 was significantly specific to IL1RAP, as no binding signal of IL1RAP was detected on the surface of IL1RAP CRISPR-Cas9 edited cell line generated in this study. Two scFv antibodies targeting IL1RAP were isolated by phage and yeast display screening. scFv-02 could serve as a promising material for further development of IL1RAP targeted immunotherapy of leukemia and other diseases. Our work also modified the process for the selection of scFv against targeted antigen by stepwise using phage display, yeast display and mammalian cell display techniques.
Introduction
Acute myeloid leukemia (AML) as well as other types of leukemia is a hematologic malignancy associated with a poor survival rate. Although cytotoxic chemotherapy is still the main modalities for treatment of leukemia, immunotherapy especially chimeric antigen receptor (CAR)-engineered T-cell (CAR-T cells) technology has turned to be a powerful strategy, which satisfies the pressing need of increased efficacy and decreased toxicity. Application of immunotherapy requires high quality of antibodies that specifically targeting cancer associated antigen.
The interleukin 1 receptor accessory protein (IL1RAP) is one of the most promising marker antigens for immunotherapy of leukemia. IL1RAP was initially known as a crucial component of the IL-1 signaling pathway [Citation1], and later expanded of several central inflammatory pathways including IL-33, IL-36G, stem cell factor (SCF) and others [Citation2–4]. IL1RAP activates pro-inflammatory signaling and promotes proliferation of candidate leukemia stem cells. IL1RAP exhibits consistently high expression in myeloid malignancies including both AML and chronic myeloid leukemia (CML), but not in hematopoietic stem cells [Citation5, Citation6]. Although its multiple functions are not completely clarified, apparent indispensability of IL1RAP for the viability of human cells points it as a promising target for myeloid malignancies therapy.
IL1RAP has recently emerged as an attractive antibody based therapeutic target in cancer. Anti-IL1RAP antibodies have been shown to significantly delay AML pathogenesis with no apparent effect on normal hematopoiesis [Citation7] and also kill primary cells from AML patients [Citation8]. Humanized anti-IL1RAP antibodies was also used for IL1RAP induced NFkB activity [Citation9]. CAR T cells targeting IL1RAP exhibited cytotoxicity against both leukemic stem cells and monocytes, without perturbing healthy hematopoietic cells [Citation10]. While as, widely application of immunotherapy targeting IL1RAP asks for an increasing number of specific antibodies.
Single-chain variable fragment (scFv) is an efficient substitute for whole antibody with reduced complexity and superior penetration ability in tissues as well as simple mode of generation. Here, we took advantage of phage display technology and screened for scFvs against IL1RAP from phage-displayed humanized scFv library. The high-affinity sublibrary was eluted out and amplified, followed by further screening through yeast display selection. Two pieces of scFvs were selected and expressed using the mammalian cell secreted expression system. scFv protein containing Fc segment was purified and the affinity of scFv to the IL1RAP on the cell surface was assessed. Binding affinity of the selected scFc to cells expressing IL1RAP was significantly higher than the affinity to IL1RAP knock-out cell surface. Our work modified the process for identification of scFv against targeted antigen by using phage display, yeast display and mammalian cell display techniques. The selected scFvs are therefore a basis for IL1RAP targeted immunotherapy of leukemia and other diseases.
Materials and methods
Phage library construction and panning
The phage-display human antibody library was based on the fusion of the scFv format to the N terminus of pIX. Total RNA was prepared from 25 different samples of human peripheral blood lymphocytes by using an RNA Purification kit (Invitrogen, Thermo Fisher Scientific Inc., USA). First strand cDNA was synthesized from total RNA by using a First-Strand cDNA Synthesis kit (ThermoFisher Scientific Inc., USA) with random hexamers. Primers and procedure to amplify the VH and VL genes from the cDNA were following the protocol published previously through two subsequent PCRs [Citation11]. The library was subjected to two rounds of panning according to standard protocol with minor modification [Citation11]. The panning involves the addition of the antibody library to immobilized antigen, washing out of unbound phage antibody clones and elution of the bound phages and amplification of selected clones by infecting Omnimax Escherichia coli with the eluted phage. The amplified phage is then subjected to the next round of panning. Human IL1RAP (Sino Biological; 10121-H08H) was biotinylated using a Sulfo-NHS-SS-Biotinylation kit (ThermoFisher Scientific Inc., USA), according to the manufacturer’s instructions.
Polyclonal phage ELISA
Polyclonal phage ELISA was used on the phage selected from each round of panning to check whether selected scFv population was specifically enriched for targeting IL1RAP. Briefly, plate wells coated with IL1RAP antigen were added with indicated phage supernatant, and secondary antibodies of HRP/anti-M13 (1:5000) (GE Healthcare; 27-9421-01) were added for detection with substrate ABTS (Roche; 11219400). The absorbance was measured at 405 nm.
Yeast library construction and screening
Enriched IL1RAP scFv candidate fragments were released from selected phage sub-library with SfiI digestion. The scFv sequences were then subcloned into pCTcon2 to construct the yeast display sub-library. Yeast display library plasmids were transformed into EBY100 and incubated with biotinylated IL1RAP prior to sorting by flow cytometry. Library generation and sorting were performed as described in the published protocol [Citation12]. Staining and flow cytometry analysis was carried out as described in our previous work [Citation13]. Anti-Myc (chicken IgY; Invitrogen A21281), secondary goat anti-chicken IgY-FITC (Thermo Fisher Scientific; PA1-28794), NeutrAvidin 633 (Thermo Fisher Scientific; 22844) and SA-PE (Thermo Fisher Scientific; S866) were used for flow cytometry screening and analysis.
Expression and purification of selected scFv-Fc variants
The IL1RAP scFv-02 and scFv-61 fragments (Supplemental Table S1) selected from yeast screening were released by SfiI digestion and subcoloned into pFuse to construct scFv with Fc tag. Opti-MEM I (Thermo Fisher Scientific; 51985049) was used to transform plasmids into 293F cells for protein expression. Supernatant of 293F cells was prepared on day 5 after transfection with the indicated plasmids. Protein A column was used to purify the secreted Fc tagged scFv antibody by AKTA purifier 100.
CRISPR-Cas9 mediated editing of IL1RAP gene
Human PE-conjugated anti ILRAP antibody (R&D Systems; FAB676P) was purchased to detect the expression of ILRAP on the surface 293F cells. sgRNAs at the target near the transmembrane region of IL1RAP (sequences in Supplemental Table S1) were designed and integrated into pGL-U6-sgRNA-puro plasmids according to Zhang Feng’s laboratory protocol [Citation14]. Constructs were individually transiently transformed into 293T cells for editing, and the efficiency was tested using T7 Endonuclease I (NEB; M0302) according to the standard procedure using a pair of checking primers (IL1RAP-CHK-F and IL1RAP-CHK-R; Supplementary Table S1). The targeted sequences #2 and #3 were integrated into pLenti-CRISPR-V2 and the constructs were packaged with lentivirus and transformed into 293F cells for editing.
Results and discussion
Screening of phage-displayed scFvs for clones population targeting IL1RAP
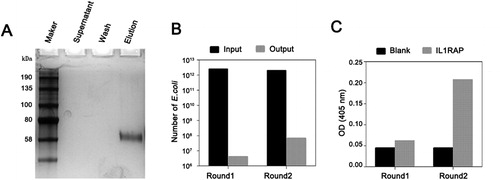
Human IL1RAP proteins were biotinylated and enriched by magnetic beads coated with streptavidin. No protein was detected in supernatant and wash go-through solution, and IL1RAP proteins were completely biotinylated and eluted as shown by silver staining (). Biotinylated IL1RAP protein was applied to screen phage-display humanized scFv as an antigen target. Two rounds of panning were carried out, in which we decreased the concentration of the biotinylated IL1RAP by 10-fold and increased washing numbers for the second-round panning. By enhancing the stringency of the second-round panning process, the enriched output specific phages increased dramatically in round 2 (). Specificity of output anti-IL1RAP scFv phage clones after each round of panning was measured by polyclonal phage ELISA. As the results revealed in , a significantly higher binding ability to the target was shown in the second-round panning compared to round 1 panning, indicating specific phage clones for IL1RAP were successfully selected from these two rounds of panning. These results demonstrated the effective enrichment of phages with the capacity to bind to IL1RAP antigen.
Figure 1. Enrichment of anti-IL1RAP scFvs by phage panning technique. (A) Silver staining for biotinylated IL1RAP protein. Note: Purchased human IL1RAP proteins were biotinylated and enriched by magnetic beads coated with streptavidin, and the supernatant was collected. After a wash, the biotinylated protein was eluted from beads. Each sample was checked by SDS-PAGE electrophoresis followed by silver staining. (B) Enrichment analysis of phage library screening for IL1RAP scFv. (C) Specific scFv enrichment analysis by ELISA. Note: IL1RAP proteins were coated and the phages population was added into the wells to detect their binding. The absorbance was read at OD405.
Selection of yeast-displayed candidate anti-IL1RAP scFvs for higher affinity and specificity
Next, we subcloned the enriched anti-IL1RAP scFv phage library to yeast display plasmids, constructing an anti-IL1RAP scFv yeast library for further screening. We then performed FITC staining to detect the Myc tag (scFv-Myc) and SA-PE staining to detect the biotinylated IL1RAP antigen, followed by flow cytometry analysis. Negative controls in the absence of antigen and irrelevant antigen were performed to confirm only IL1RAP antigen positive cell populations being gated. After the first screening round by employing 100 nmol/L antigens, pools of scFvs with high affinity for IL1RAP were able to be enriched (from 100% phage clones to 21.6% yeast population) ().
Figure 2. Selection of yeast displayed anti-IL1RAP scFv by flow sorting. (A) A yeast-display IL1RAP scFv library enriched for scFv binding to 100 nmol/L of biotinylated IL1RAP protein by first round of flow sorting. (B) The second round of selection for scFv binding to 50 nmol/L of biotinylated IL1RAP protein. (C) The third round of selection for scFv binding to 10 nmol/L of biotinylated IL1RAP protein. Note: IL1RAP binding to yeast-display scFv was measured by staining with biotinylated IL1RAP protein and anti-c-Myc antibody detected by PE-labeled streptavidin (or APC-labeled neutravidin) and FITC-labeled secondary antibody (c, f, i). As negative controls, yeast cells were not stained (a, d, g) and incubate without antigen (b, h) or biotinylated Her2 protein (e).
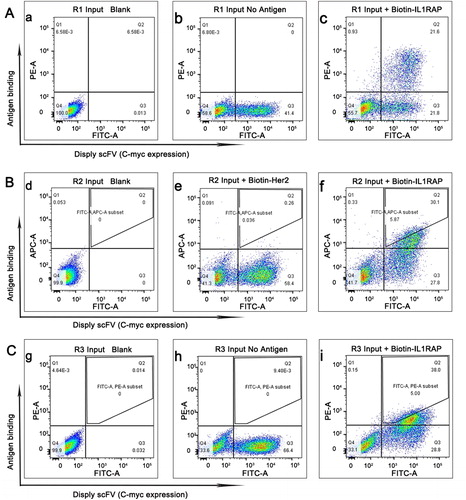
In the second screening round, we enhanced the stringency by employing 50 nmol/L antigens for selection of scFv clones with higher affinity and specificity. In addition, we replaced PE-labeled streptavidin by APC-labeled neutravidin to get rid of the cross-reactive binding of streptavidin to the yeast. As a control, we also used an irrelevant antigen biotinylated Her2 to test the specificity of the enriched anti-IL1RAP scFv clones. The results demonstrated very specific enrichment of yeast displaying scFv targeting IL1RAP (). For a better affinity and specificity, we only sorted and collected a subset of desired scFv in Q2 zone ().
The collected clones were amplified and applied to a further selection with a stricter setting of very low concentration of IL1RAP. The concentration of antigen used in the third round was only 10 nmol/L, 1/10 of that used in the initial selection. The enriched scFv should be of best affinity and specificity after three rounds of selection with stepwise increased screening stringency. Moreover, we collected only the desired population from the positive Q2 zone in the third selection output clones (). These output clones were collected for further analysis.
Sequence analysis and characterization of obtained anti-IL1RAP scFvs
100 individual clones were picked randomly from the yeast scFv selection output population and screened for sequence analysis to isolate the accurate functional framework of the heavy and light chain. These errors included clones were excluded out after sequence analysis and the sequence analysis revealed 92 of 100 clones had an acceptable sequence. Among them, precisely only two pieces of fragments were hit after the whole screening, in which scFv-02 appeared 88 times and the rest were scFv-61. NCBI-BLAST tools and Clustal version 2.1 software were used to determine the sequence alignment and homologous sequence analysis with scFv-02 and scFv-61 (). Sequence analysis of the isolated binders of scFv-02 and scFv-61 revealed clustering of identical heavy chain CDR1 and CDR2 regions, but still high diversity in light chain regions ().
Figure 3. Sequence analysis and binding affinity test of selected positive scFvs. (A) Sequence alignment of scFv-02 and scFv-61. Alignments indicating highly conserved sequences are shown in colors. The CDRs of the variable domains are indicated. (B) Structural configuration of scFv-02 and scFv-61 by protein modeling software Swiss-model. (C) Analysis of anti-IL1RAP scFv binding by flow cytometry. Note: IL1RAP binding to yeast-display scFv was measured by staining with 5 nmol/L of biotinylated IL1RAP protein. FITC staining to detect the Myc tag (scFv-Myc) and SA-PE staining to detect the biotinylated IL1RAP antigen.
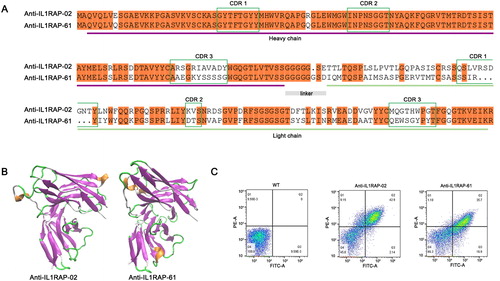
Interestingly, both scFvs against IL1RAP contained a short sequence of Gly4Ser in linker peptide instead of a Gly4Ser repeat. Previous study indicated that the linker peptide between variable domains of heavy (VH) and light (VL) chains containing (Gly4Ser)n is one of the important factors that influence the binding activity and specificity of scFv against the target antigen [Citation15, Citation16]. Varying the length of the linker peptide also affected scFv characterization. Shortening the linker of the VH-VL oriented scFv may be attractive in some important properties [Citation16–18]. Our work obtained two scFvs against IL1RAP that were both unexpectedly mutated to a short sequence of linker peptide (). Structural configuration of scFv-02 and scFv-61 modeled by protein modeling software Swiss-model indicated classical conformations for antigen binding, including VH and VL variable regions and the linker between VH and VL ().
To further analyze scFv-02 and scFv-61, we re-transformed the corresponding plasmids to fresh EBY100 yeast cells individually and examined their binding activity to biotinylated IL1RAP antigen. Flow cytometry showed that yeast cells displaying either scFv-02 or scFv-61 were significantly positive for IL1RAP binding, although low concentration of antigen (5 nmol/L) was given for the test (). Compared with scFv-61, clone scFv-02 exhibited higher binding activity which is compatible with their hit frequency in the screening.
Binding specificity of selected scFvs against IL1RAP on 293F cell surface
To test the ability of anti-IL1RAP scFv to recognize the target on the surface of human cells, we took advantage of human 293F cell lines for binding test. First, we confirmed the expression of IL1RAP on the surface of 293F cells by flow cytometry using purchased PE-labeled anti-IL1RAP antibody (). For direct analysis using an IL1RAP null mutant as a negative control, we mutated IL1RAP gene in 293F cells by using CRISPR-Cas9 editing to create IL1RAP knock out (KO) 293F cells. We designed three sgRNAs targeting different sites of IL1RAP gene and checked the efficiency by T7EI assay. As shown in , all sgRNAs selected were successful for sgRNA guided editing in IL1RAP gene. We selected sgRNA-2 and sgRNA-3 as a mixture for transient transforming 293F cells and generated IL1RAP-KO 293F cells as a negative control for scFv binding assay.
Figure 4. Binding specificity of selected scFvs with IL1RAP antigen on 293F cell surface. (A) FACS analysis of endogenous expression of IL1RAP on 293F cell surface. Purchased PE-labeled anti-IL1RAP antibodies were used to detect the endogenous expression of IL1RAP on 293F cell surface. (B) T7EI assay to confirm the efficiency of CRISPR-Cas9 mediated IL1RAP editing. (C) Cell binding assay to verify the specificity of scFv-02 and scFv-61 to IL1RAP antigen on 293F cell surface.
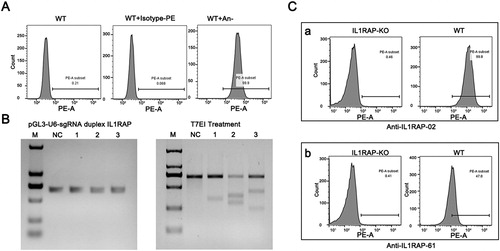
On the other hand, we expressed the selected scFvs by heterologous expression of them as Fc fusion proteins in a mammalian Expi-293F system and after purification protein yields of around 20 mg/L cell culture were obtained. To test the ability of selected anti-IL1RAP scFvs to recognize target on mammalian 293F cell surface, we incubated 293F or IL1RAP-KO 293F cells with purified scFv-Fc proteins (02 and 61) followed by staining and flow cytometry analysis. As expected, both anti-IL1RAP scFvs bound to the wild-type 293F cells but not to IL1RAP null mutants (). scFv-02 purified from mammalian expression system also showed a significantly higher binding ability to 293F cell surface than scFv-61 (). Taken together, these results strongly supported that both scFvs recognize IL1RAP antigen very well on the surface of human cells, which provides a promising material for further development of IL1RAP targeted immunotherapy of leukemia and other diseases.
Conclusions
Here, we are successful in isolating two highly specific scFvs against IL1RAP. We also provide a simple and rapid method for the quick identification of a scFv targeting IL1RAP. We generate IL1RAP null cells by using CRISPR-Cas9 editing for direct analysis of the selected scFv binding specificity to the target. Our study also demonstrates the superiority of cell panning strategy via stepwise use of phage and yeast display for selection of scFv targeting IL1RAP from a human library. Further characterization of the isolated scFv antibodies is necessary to fully elucidate their potential in cancer diagnosis and therapy.
Supplemental Material
Download PDF (409.2 KB)Disclosure statement
The authors declare no conflict of interest.
Additional information
Funding
References
- Korherr C, Hofmeister R, Wesche H, et al. A critical role for interleukin-1 receptor accessory protein in interleukin-1 signaling. Eur J Immunol. 1997;27(1):262–267.
- Palmer G, Lipsky BP, Smithgall MD, et al. The IL-1 receptor accessory protein (AcP) is required for IL-33 signaling and soluble AcP enhances the ability of soluble ST2 to inhibit IL-33. Cytokine 2008;42(3):358–364.
- Drube S, Heink S, Walter S, et al. The receptor tyrosine kinase c-Kit controls IL-33 receptor signaling in mast cells. Blood. 2010;115(19):3899–3906.
- Shastri A, Will B, Steidl U, et al. Stem and progenitor cell alterations in myelodysplastic syndromes. Blood. 2017;129(12):1586–1594.
- Barreyro L, Will B, Bartholdy B, et al. Overexpression of IL-1 receptor accessory protein in stem and progenitor cells and outcome correlation in AML and MDS. Blood. 2012;120(6):1290–1298.
- Järås M, Johnels P, Hansen N, et al. Isolation and killing of candidate chronic myeloid leukemia stem cells by antibody targeting of IL-1 receptor accessory protein. Proc Natl Acad Sci USA. 2010;107:16280–16285.
- Mitchell K, Barreyro L, Todorova TI, et al. IL1RAP potentiates multiple oncogenic signaling pathways in AML. J Exp Med. 2018;215(6):1709–1727.
- Askmyr M, Ågerstam H, Hansen N, et al. Selective killing of candidate AML stem cells by antibody targeting of IL1RAP. Blood. 2013;121(18):3709–3713.
- Fisher S, Brandt M. 2017. Humanized anti-IL-1R3 antibodies. Patent, WO 2017/191325 A9.
- Warda W, Larosa F, Neto Da Rocha M, et al. CML hematopoietic stem cells expressing IL-1RAP can be targeted by chimeric antigen receptor (CAR)-engineered T cells. Cancer Res. 2019;79(3):663–675.
- Gao C, Mao S, Kaufmann G, et al. A method for the generation of combinatorial antibody libraries using pIX phage display. Proc Natl Acad Sci USA. 2002;99(20):12612–12616.
- Van Deventer JA, Wittrup KD. Yeast surface display for antibody isolation: Library construction, library screening, and affinity maturation. Methods Mol Biol. 2014;1131:151–181.
- Jia Y, Ren P, Duan S, et al. An optimized yeast display strategy for efficient scFv antibody selection using ribosomal skipping system and thermo resistant yeast. Biotechnol Lett. 2019;41(8–9):1067–1076.
- Joung J, Konermann S, Gootenberg JS, et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc. 2017;12(4):828–863.
- Yusakul G, Sakamoto S, Pongkitwitoon B, et al. Effect of linker length between variable domains of single chain variable fragment antibody against daidzin on its reactivity. Biosci Biotechnol Biochem. 2016;80(7):1306–1312.
- Paudel MK, Sakamoto S, Van Huy L, et al. The effect of varying the peptide linker length in a single chain variable fragment antibody against wogonin glucuronide. J Biotechnol. 2017;251:47–52.
- Dolezal O, De Gori R, Walter M, et al. Single-chain Fv multimers of the anti-neuraminidase antibody NC10: The residue at position 15 in the V(L) domain of the scFv-0 (V(L)-V(H)) molecule is primarily responsible for formation of a tetramer-trimer equilibrium. Protein Eng. 2003;16(1):47–56.
- Arndt MA, Krauss J, Rybak SM. Antigen binding and stability properties of non-covalently linked anti-CD22 single-chain Fv dimers. Febs Lett. 2004;578(3):257–261.
