Abstract
Ardisia gigantifolia is a medicinal plant; its herbal medicinal product has played important roles in the ways of life of people in China. However, commercial products of this herbal medicine are frequently adulterated with Rhododendron molle root and Clerodendrum cyrtophyllum stem. In fact, R. molle has hepatotoxic effects which could pose health issues to the end user. In this study, we applied DNA barcoding coupled with high resolution melting (Bar-HRM) to distinguish A. gigantifolia and adulterant products. Two DNA barcode loci (ITS2 and psbA-trnH) were chosen for Bar-HRM analysis. We found that ITS2 and psbA-trnH regions contain some inter-species variability, which showed enough discrimination capability for Bar-HRM analysis. We applied the Bar-HRM method to test the method, and found that some products labelled as “A. gigantifolia” collected from local markets in China were identified to contain C. cyrtophyllum. Our study demonstrated that DNA barcoding coupled with high resolution melting can be used as a routine test to guarantee the quality of A. gigantifolia herbal products.
Introduction
Ardisia gigantifolia, an endangered medicinal plant belonging to the genus Ardisia, is distributed only in Guangxi, Guangdong, Fujian and Jiangxi provinces, China [Citation1]. The dry root of this plant is used as an important traditional Chinese medicine to treat rheumatoid arthritis and inflammatory diseases in China for thousands of years as evidenced by ancient records [Citation2]. It is also widely used as a remedy for bruises, sprains, blood stasis, tumors and chronic ulcer diseases. Recently, various types of bioactive ingredients have been isolated from this medicinal plant, including quinones, coumarins, saponins and so on. Furthermore, compounds isolated from this species have a range of bioactivities, for example anti-HIV, anti-inflammation, anti-tumor, anti-oxidation properties and antivirus activity [Citation3]. However, with the market demand increasing and wild resources decreasing, the population of A. gigantifolia has been destroyed at a phenomenal rate, and regarded as a rare medicinal plant [Citation4]. Because of its high market demand, the price of A. gigantifolia is more expensive than other related medicinal plants in markets. The high price has led to substantially adulterated samples by the intentional substitution with other cheaper plants. Market surveys identified Rhododendron molle root and Clerodendrum cyrtophyllum stem as the main medicinal materials used to adulterate commercial products of A. gigantifolia [Citation5]. Recently, evidence that R. molle has hepatotoxic effects was reported [Citation6–7]. However, identifying medicinal materials by morphological characters is complicated because they have similar morphology [Citation5]. It is more difficult to discern medicinal materials when they are dried, cut, or pulverized. Therefore, the accurate identification of A. gigantifolia is inportant for authenticity and quality of Chinese medicinal herb, as well as safety and health for consumers.
With the development of molecular biology, DNA-based methods have been used to identify medicinal plants more accurately. Among these methods, DNA barcoding is a useful tool and has been widely applied to identify original species to guarantee food safety and study biodiversity [Citation8–10]. In 2010, ITS2 was suggested as a universal barcoding locus for most medicinal plants [Citation11]. Since then, the ITS2 locus has been widely used for medicinal plants identification [Citation12–15]. Meanwhile, a chloroplast genome region psbA-trnH has also been proposed as a complementary marker for barcoding medicinal plants [Citation16]. These studies demonstrated that DNA barcoding is conducive to addressing the difficulties of identifying herbal materials and provide a new taxonomic method for herbal plants.
High resolution melting (HRM) is a new method based on PCR. This technique could distinguish different genotypes by the different shapes of whole melting transitions (Tm) for different real-time PCR products. When the temperature increases, the DNA duplex is denatured, the intercalating DNA dye releases from the DNA double helix and the fluorescence changes. This technology is used for sequence matching, mutation scanning and genotyping polymorphisms, and is increasingly applied in medical research, such as cancer testing [Citation17–18]. Recently, it is also used for authentication of raw materials and products of Chinese medicinal herbs, as well as for exact quantification of the proportion of adulterants in herbal products [Citation18–19].
This study aimed to establish a combined method to distinguish the commercial A. gigantifolia product and its adulterants accurately using DNA barcoding coupled with HRM analysis. Two DNA barcode sequences (ITS2 and psbA-trnH) were used to examine their effectiveness as barcodes for Bar-HRM in the identification of A. gigantifolia. Finally, Bar-HRM was applied to evaluate different commercial herbal medicines labelled as “A. gigantifolia” species collected from the local market.
Materials and methods
Plant material
Forty-three authenticated samples which were collected from southern China were used to validate the Bar-HRM assay. These contained nineteen A. gigantifolia samples, twenty C. cyrtophyllum, and four R. molle samples. The morphological identification of these samples was completed by Shizhong Mao at Guangxi Institute of Botany, Chinese Academy of Sciences. Each voucher specimen was kept in the Herbarium of Guangxi Institute of Botany, Chinese Academy of Sciences. In addition to voucher samples, twelve commercial products of A. gigantifolia species were bought from local markets in China.
DNA isolation and HRM-PCR amplification
The plant materials were frozen with liquid nitrogen and then ground into a powder. For each sample, genomic DNA was extracted from 50 mg of powder by the Plant Genomic DNA Kit (Tiangen Biotech Co.). The DNA concentrations of samples were measured by a spectrophotometer (Qubit 3.0, Invitrogen Co., USA), and then the samples were diluted to 50 ng/μL for the next step.
The ITS2 region and the psbA-trnH region were selected for identity analysis because they have low intra-specific sequence variation but have sufficient variable sites to distinguish most closely related species [Citation12,Citation13,Citation16]. Universal primers from these two regions (psbAF: 5′-GTT ATG CAT GAA CGT AAT GCT C-3′, trnHR: 5′-CGC GCA TGG ATT CAC AAT CC-3′ and ITS2F: 5′-ATG CGA TAC TTG GTG TGA AT-3′, ITS3R: 5′-GAC GCT TCT CCA GAC TAC AAT-3′) were used for HRM-PCR. The HRM-PCR reactions using both regions and the subsequent data analysis were carried out using to the method described by Li et al [Citation20]. The only difference was that the annealing temperature for the ITS2 region was 56 °C, while for the psbA-trnH region was 55 °C.
PCR product sequencing and species identification
After HRM analysis, all PCR products were directly sequenced using the automated ABI 3730 XL sequencer (Applied Biosystems). Each sequence was assembled using CodonCode Aligner (CodonCode, USA), and then aligned using MEGA5.1 software [Citation21]. Finally, all the sequences were submitted to an online DNA barcoding database of herbal materials identification (http://www.tcmbarcode.cn). This database provides a system for herbal species identification, and also provides barcoding sequences for their closely related species, adulterants and substitutes [Citation22]. The Minimum Standards of Reporting Checklist contains details of the experimental design, and statistics, and resources used in this study.
Results
Analysis of ITS2 and psbA-trnH sequences
The sequences of ITS and psbA-trnH loci of A. gigantifolia and its adulterants were submitted to DNA Barcoding System for Identifying Herbal Medicine (http://www. tcmbarcode.cn) for species discrimination. The results showed that herbal materials could be identified using this system (). The success rates for PCR amplification and sequencing of all the samples were 100% for the two barcodes. In total, 86 sequences for 43 individual plants were analyzed in this study.
Table 1. Original information of plant materials used in this study.
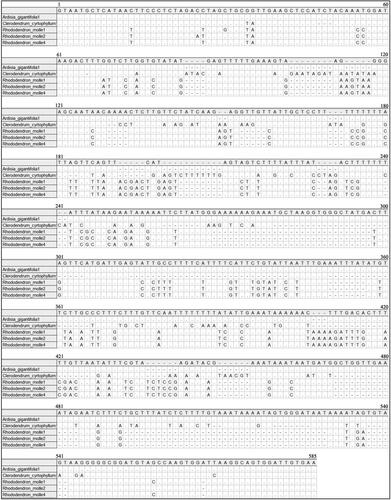
The length of the ITS2 sequences varied from 217 bp to 230 bp. For the ITS2 matrix, the intraspecific distances ranged from 0 to 0.48% and the interspecific distances ranged from 44.75% to 59.04%. The length of psbA-trnH sequences varied from 450 bp to 528 bp. For the psbA-trnH matrix, the intraspecific distances ranged from 0 to 0.87% and the interspecific distances ranged from 19.07% to 28.61%). The results showed that the two barcodes have high interspecific divergence and were suitable for further HRM analysis ( and ). The information of sequences (ITS2 and psbA-trnH) are available at https://figshare.com/s/e124acf847252f93d9b4.
Figure 1. Alignment of ITS2 locus of sequencing products of three herbal species, alignment information can be accessed at https://figshare.com/s/e124acf847252f93d9b4.
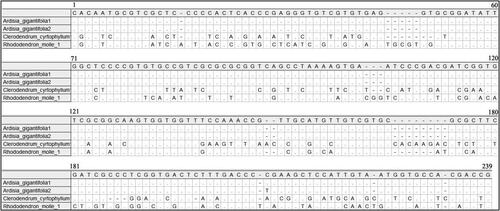
Figure 2. Alignment of psbA-trnH locus of sequencing products of three herbal species, alignment information can be accessed at https://figshare.com/s/e124acf847252f93d9b4.
HRM analysis based on ITS2 and psbA-trnH sequences
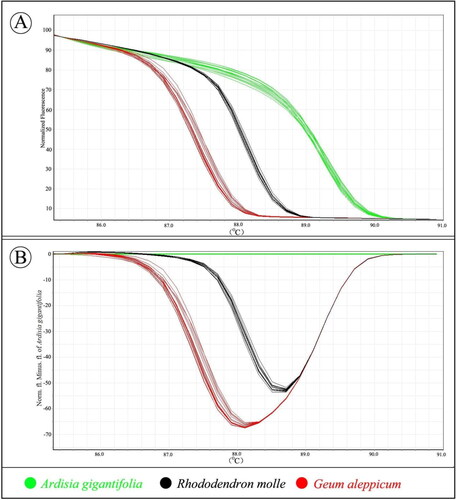
Two different curves were plotted to assess the melting characteristics of the ITS2 amplicons of all voucher samples (). The three species were easily distinguished by different melting profiles using barcode marker ITS2 (). In the difference-plot melt curves (), improved visualization and separation of variant melting curves of each species were displayed. By assigning A. gigantifolia as the reference genotype, three species were successfully genotyped. Finally, all the examined species were shown indeed to be different, while the mean A. gigantifolia curve was used as the baseline.
Figure 3. High resolution melting analysis of a fragment of ITS2. (A) Curves of three herbal species in a normalized melting curve graph. (B) Difference graph of the normalized melting curves using A. gigantifolia as baseline. Information can be accessed at https://figshare.com/s/e124acf847252f93d9b4.
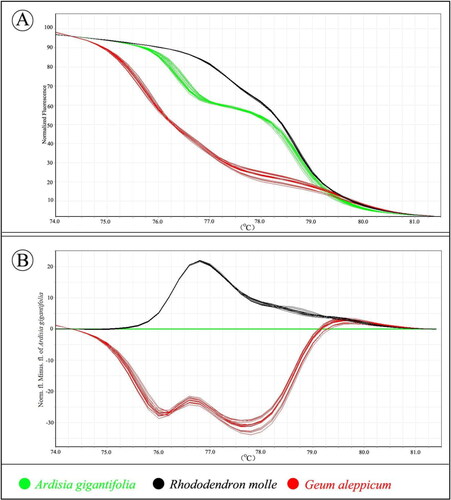
Analysis of the normalized HRM curves based on the barcode locus psbA-trnH is shown in . Three species could be successfully distinguished because of their different melting profile (). Furthermore, using the mean A. gigantifolia curve as the baseline, closer examination of the HRM difference curve revealed part of the curve sitting outside the 90% CI curve, and the result suggested that the three examined species were indeed different (). DNA sequencing also confirmed the results obtained by HRM. These results all demonstrated that psbA-trnH was a sufficient marker to distinguish A. gigantifolia from its toxic adulterants.
Figure 4. High resolution melting analysis of a fragment of psbA-trnH. (A) Curves of three herbal species in a normalized melting curve graph. (B) Difference graph of the normalized melting curves using A. gigantifolia as baseline. Information can be accessed at https://figshare.com/s/e124acf847252f93d9b4.
Species identification of commercial products from local markets
Twelve samples of A. gigantifolia commercial herbal products were collected from Chinese local markets, and then were used to identify their accurate ingredients by Bar-HRM analysis. When using ITS2 primers, out of twelve samples, two samples labelled as A. gigantifolia were identified as C. cyrtophyllum (). Using psbA-trnH primers, the same result was obtained (). Each sequence of A. gigantifolia commercial herbal product was also analyzed to assess the results of HRM analysis. The blast results were consistent with the results by Bar-HRM ().
Figure 5. Melting profiles of commercial products based on ITS2 region. (A) Normalized fluorescence curves of high resolution melting; (B) Reference-corrected normalized fluorescence plot of HRM. Information can be accessed at https://figshare.com/s/e124acf847252f93d9b4.
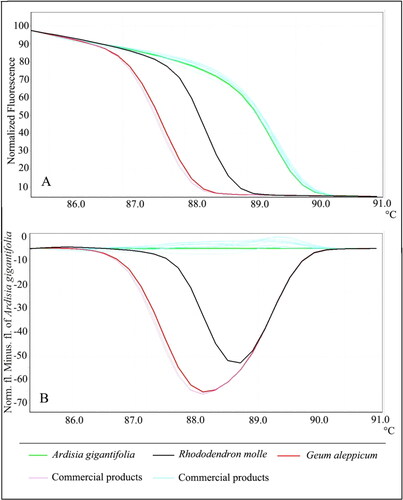
Figure 6. Melting profiles of commercial products based on psbA-trnH region. (A) Normalized fluorescence curves of high resolution melting; (B) Reference-corrected normalized fluorescence plot of HRM. Information can be accessed at https://figshare.com/s/e124acf847252f93d9b4.
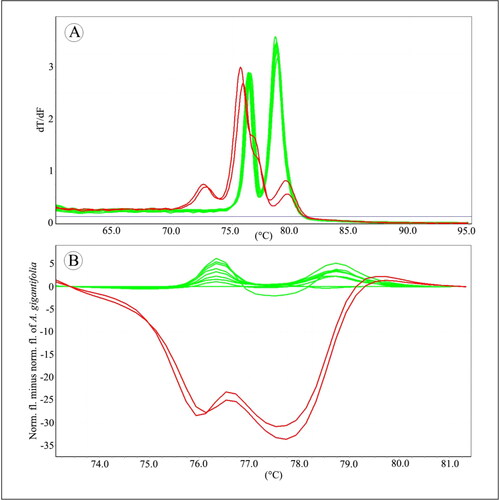
Discussion
DNA barcodes are widely used for detecting substitution and adulteration of Chinese herbal medicines. In systematic research, ITS2 has been proposed as the core DNA barcode, while psbA-trnH was proposed as a supplementary DNA barcode to authenticate medicinal plants. Many previous studies have validated the identification capabilities of ITS2 and psbA-trnH locus [Citation12–16]. The major advantages of DNA barcoding is its high sensitivity, and specificity. However, this method requires a molecular laboratory, costly equipment, and DNA sequencing machine; this inherent limitation hinders the wider implementation of DNA barcoding in developing countries. Therefore, to develop a sequencing free method that is faster and more economical than DNA barcoding is urgent. Bar-HRM is such a method and its discrimination capability is based on the length or base polymorphism of the target DNA regions. Bar-HRM could be used to identify the original species of traditional herbal products, whether a sample was a single ingredient sample or contained adulterants. Furthermore, the complete analysis of this simple and reliable technology is conducted in a closed tube system, and prevents cross-contamination risk [Citation23–25]. Previous studies showed that Bar-HRM method has a potential powerful ability in identification for some herbal medicines [Citation26–29]. However, until now, HRM analysis has not been widely utilized for identification of herbal products [Citation30–32].
In this work, two DNA barcode regions (ITS2 and psbA-trnH) were selected for the authentication of A. gigantifolia. Bar-HRM assays were developed for both ITS2 and psbA-trnH regions. The results revealed that Bar-HRM is a simple and fast approach suitable for species identification. After HRM analysis, the PCR product of each sample was directly sequenced and the results confirmed that the samples could be directly genotyped by using the melting curves. For the determination of commercial frauds, a total of twelve commercial products sold as A. gigantifolia were analyzed. Two of these products were found to contain the ingredient of C. cyrtophyllum. Although this test did not find that the A. gigantifolia commercial herbal products contain toxic R. molle species on the market, our result demonstrated that DNA barcoding coupled with high resolution melting can provide a reference for the quality control and management strategies of A. gigantifolia products, as well as applied to test other herbal products [Citation33–35].
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
All data generated during this study are available at Figshare (https://doi.org/10.6084/m9.figshare.13042928).
Additional information
Funding
References
- Feng JQ, Huang ZX, Mu LH, et al. Study on chemical constituents of rhizome of Ardisia gigantifolia. China Journal of Chinese Materia Medica. 2011;36(24):3463–3466.
- Mu LH, Zhao HX, Gong QQ, et al. Triterpenoid saponins and the antitumor activity from the rhizome of Ardisia Gigantifolia Stapf. Pharmacological Journal of Chinese PLA. 2011;27:1–6. Chinese.
- Mu LH, Wang LH, Wang YN, et al. Antiangiogenic effects of AG36, a triterpenoid saponin from Ardisia gigantifolia stapf. J Nat Med. 2020; 74(4):732–740.
- Mao SZ, Tang WX, Luo WH, et al. Medicinal plant resources of Ardisia in Guangxi and sustainable utilization. Journal of Fujian Forest Science and Technology. Chinese. 2010;37(2):119–126.
- Yu JC, Zhang LQ, Li Y, et al. Identification Ardisia gigantifolia and its adulterant Rhododendron molle. Strait Pharm J. 2011;23:49–50. Chinese.
- Klocke JA, Hu MY, Chiu SF, et al. Grayanoid diterpene insect antifeedants and insecticides from Rhododendron molle. Phytochemistry. 1991;30(6):1797–1800.
- Bernstein WB, Trotta RF, Rector JT, et al. Mechanisms for linezolid-induced anemia and thrombocytopenia. Ann Pharmacother. 2003;37(4):517–520.
- Gao T, Yao H, Song J, et al. Evaluating the feasibility of using candidate DNA barcodes in discriminating species of the large Asteraceae family. BMC Evol Biol. 2010;10(1):324
- Haye PA, Segovia NI, Vera R, et al. Authentication of commercialized crab-meat in Chile using DNA barcoding. Food Control. 2012;25(1):239–244.
- Yan LJ, Liu J, Möller M, et al. DNA barcoding of Rhododendron (Ericaceae), the largest Chinese plant genus in biodiversity hotspots of the Himalaya–Hengduan Mountains. Mol Ecol Resour. 2015;15(4):932–944.
- Chen SL, Yao H, Han JP, et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One. 2010;5(1):e8613
- Pang X, Shi L, Song J, et al. Use of the potential DNA barcode ITS2 to identify herbal materials. J Nat Med. 2013;67(3):571–575.
- Sun Z, Gao T, Yao H, et al. Identification of Lonicera japonica and its related species using the DNA barcoding method. Planta Med. 2011;77(3):301–306.
- Zomuanpuii R, Ringngheti L, Brindha S, et al. ITS2 characterization and Anopheles species identification of the subgenus Cellia. Acta Trop. 2013;125(3):309–319.
- Sharma S, Shrivastava N. Internal transcribed spacer guided multiplex PCR for species identification of Convolvulus prostratus and Evolvulus alsinoides. Acta Pharm Sin B. 2016;6(3):253–258.
- Li M, Cao H, But PP, et al. Identification of herbal medicinal materials using DNA barcodes. J Sys Evol. 2011;49(3):271–283.
- Cai S, Xu J, Shao Y, et al. Rapid identification of the Candida glabrata species complex by high-resolution melting curve analysis . J Clin Lab Anal. 2020; 34(6):e23226
- Papaioannou C, Zeliou K, Trigas P, et al. High resolution melting (HRM) genotyping in the genus Origanum: molecular identification and discrimination for authentication purposes. Biochem Genet. 2020; 58(5):725–737.
- Lagiotis G, Stavridou E, Bosmali I, et al. Detection and quantification of cashew in commercial tea products using High Resolution Melting (HRM) analysis. J Food Sci. 2020; 85(6):1629–1634.
- Li J, Song M, Xiong C, et al. Application of barcode high-resolution melting for rapid authentication of the medicinal plant Psammosilene tunicoides. Biotechnol Biotec Eq. 2016;30(4):1–7.
- Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739.
- Chen SL, Pang XH, Song JY, et al. A renaissance in herbal medicine identification: from morphology to DNA. Biotechnol Adv. 2014;32(7):1237–1244.
- Ganopoulos I, Sakaridis I, Argiriou A, et al. A novel closed-tube method based on high resolution melting (HRM) analysis for authenticity testing and quantitative detection in Greek PDO Feta cheese. Food Chem. 2013;141(2):835–840.
- Ganopoulos I, Bazakos C, Madesis P, et al. Barcode DNA high-resolution melting (Bar-HRM) analysis as a novel close-tubed and accurate tool for olive oil forensic use. J Sci Food Agric. 2013;93(9):2281–2286.
- Ioannis G, Panagiotis M, Antonios Z, et al. High-resolution melting analysis allowed fast and accurate closed-tube genotyping of Fusarium oxysporum formae speciales complex. Fems Microbiol Lett. 2012;334(1):16–21.
- Buddhachat K, Osathanunkul M, Madesis P, et al. Authenticity analyses of Phyllanthus amarus using barcoding coupled with HRM analysis to control its quality for medicinal plant product. Gene. 2015;573(1):84–90.
- Kalivas A, Ganopoulos I, Xanthopoulou A, et al. DNA barcode ITS2 coupled with high resolution melting (HRM) analysis for taxonomic identification of Sideritis species growing in Greece. Mol Biol Rep. 2014;41(8):5147–5155.
- Xanthopoulou A, Ganopoulos I, Kalivas A, et al. Multiplex HRM analysis as a tool for rapid molecular authentication of nine herbal teas. Food Control. 2016;60:113–116.
- Duan BZ, Wang YP, Fang HL, et al. Authenticity analyses of Rhizoma Paridis using barcoding coupled with high resolution melting (Bar-HRM) analysis to control its quality for medicinal plant product. Chin Med. 2018;13(1):8
- Osathanunkul M, Madesis P. Bar-HRM: a reliable and fast method for species identification of ginseng (Panax ginseng, Panax notoginseng, Talinum paniculatum and Phytolacca Americana). Peer J. 2019; 7(3):e7660
- Chen YQ, Wang B, Wang P, et al. Rapid identification of common medicinal snakes and their adulterants using the Bar-HRM analysis method. Mitochondrial DNA A DNA Mapp Seq Anal. 2019; 30(2):367–374.
- Liu YY, Fang HL, Li XW, et al. Identification of Gentiana rhodantha and its adulterants by DNA barcoding coupled with high resolution melting analysis. Chin Pharm J. 2019; 054(009):687–692. Chinese.
- Xiong C, Hu Z, Tu Y, et al. ITS2 barcoding DNA region combined with high resolution melting (HRM) analysis of Hyoscyami Semen, the mature seed of Hyoscyamus niger. Chin J Nat Medicines. 2016;14(12):898–0903.
- Song M, Li J, Xiong C, et al. Applying high-resolution melting (HRM) technology to identify five commonly used Artemisia species. Sci Rep. 2016;6:34133
- Yue L, Li X, Yi Z, et al. DNA barcoding based identification of Hippophae species and authentication of commercial products by high resolution melting analysis. Food Chem. 2018;242:62–67.
