Abstract
Common bean is an important legume crop having high quality protein, micronutrients, vitamins and antioxidants, which makes it a “grain of hope” for poor communities. Hence, a good number of breeding activities have been performed on the improvement of various key traits for years. However, recent advancements in molecular markers, sequencing technologies and the completion of the common bean genome sequence have opened numerous opportunities for fine mapping and gene characterization. The availability of these tools together with investigations of quantitative trait loci (QTL) and candidate genes for key traits such as morpho-agronomic, iron and zinc contents, cooking and quality traits, antioxidant activity, biotic and abiotic stresses pave the way to the development of new strategies for common bean genetic improvement. As a food source, it can contribute to the reduction of food scarcity worldwide in the coming years. Therefore, it is very important to take synergic efforts to integrate common bean genetic and genomic resources in breeding activities to ensure food security and contribute significantly to improved livelihoods in developing countries. Moreover, Kompetitive allele specific PCR (KASP) and CRISPR-Cas9 should be used to develop climate resilience common bean varieties. Here, we provide an overview of the evolution of common bean research by highlighting the past and recent advances in genomics, transgenics, transcriptomics and proteomics and also critically discuss the future prospects for further genetic improvement and better expansion of this crop.
Introduction
With the rapid increase in the world’s population, it is estimated that 70% more food will be required to meet the food demand for the predicted population of 9.6 billion people by 2050, and this demand will be mainly from developing countries and Africa [Citation1]. To overcome this challenge, there is a need to increase the production of all crops all over the world, especially in Africa [Citation2]. Legumes are the major source of proteins and minerals and an important pillar of agricultural production systems, playing a vital role in the human diet and farming systems of developing countries [Citation3,Citation4].
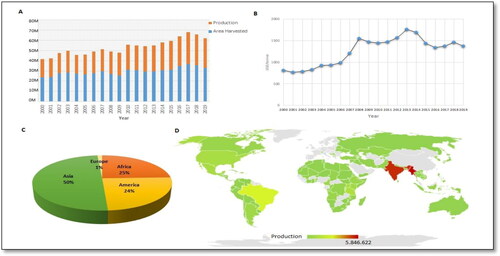
Common bean (Phaseolus vulgaris L.) is an important grain legume crop and mostly used worldwide for its pods and edible seeds [Citation5–9]. It is an important source of protein and provides 15% of protein and fulfills 30% of caloric requirements of the world’s population [Citation10]. There are 50 Phaseolus species; however, only five species (P. vulgaris, P. lunatus, P. coccineus, P. acutifolius, and P. polyanthus) have been mainly used as human food. Among these five species, common bean (P. vulgaris) is most widely used for human consumption [Citation11]. The common bean is an annual herbaceous plant with great diversity in its growth habit, for instance it may be a determinate/indeterminate bush or a climber to semi climber vine [Citation12]. According to FAO, common bean global harvested area was 33.1 million ha and production was 28.9 million tons in 2019 [Citation13]. Asia shares 50% of the global production of common bean (), and Myanmar, India, Brazil, China, America were the top five dry bean producing countries in the world in 2000–2019 [Citation13].
Figure 1. Common bean production in the world. (A) Evolution of common bean seed production and area under cultivation from 2000 to 2019. (B) Evolution of sale prices of common bean seed from 2000 to 2019. (C) Production share of common bean seed by continent in 2019. (D) Map of production quantities of common bean seed by country in 2019 (Source: Food and Agriculture Organization Statistical Databases was used to develop this figure (FAOSTAT) 2020).
Relevance of common bean research
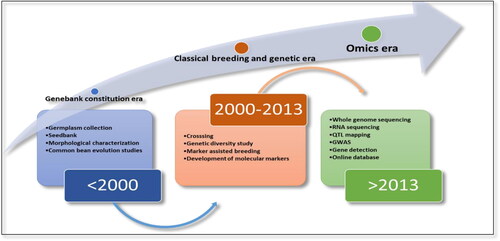
Common bean is a true diploid with a genome size of 587 Mb [Citation14]. It is considered a model crop for comparative genomics studies in different legumes due to its phylogenetic position in the Phaseoloids [Citation15]. It shows very close relatedness to papilionid legumes like soybean (Glycine max), pigeon pea (Vigna radiata) and cowpea (Vigna unguiculata) [Citation16]. Previous studies showed that the divergence of soybean and common bean occurred ∼19.2 million years ago; however, they shared a whole-genome duplication event ∼56.5 million years ago [Citation14, Citation17]. Moreover, most segments of any single common bean linkage group showed a higher level of resemblance to two soybean chromosomes on the basis of synteny analysis [Citation18]. Due to its small genome size, it can serve as a model crop to understand the genomics of higher genome size crops, like soybean, with a genome size ∼1,100 Mbp [Citation10]. Previously, common bean was considered an “orphan crop”. However, this crop entered the modern era crop after the successful genome sequencing in 2014 [Citation14]. Historical advancements in common bean are shown in .
Common bean origin and domestication
Mesoamerica is the main region of origin, where maximum numbers of Phaseolus species are geographically distributed, and it is assumed that the Phaseolus genus originated between 4 to 6 million years ago [Citation19] in this region [Citation12, Citation19]. Having originated in Mesoamerica, what is now Mexico [Citation20], wild forms of common bean were mainly distributed from northwestern Argentina to northern Mexico and resulted in the formation of unique gene pools (), namely the Andean, Mesoamerica and Peru-Ecuador [Citation21,Citation22].
Figure 3. Representation of common bean gene pool (The figure was adapted from Google Earth (https://earth.google.com/web/) and colored by authors).
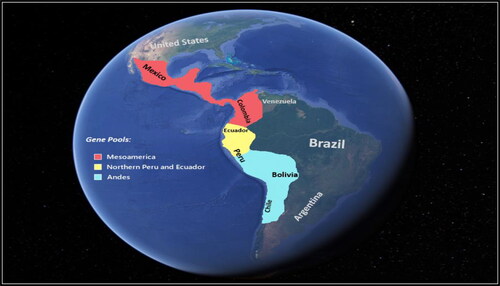
Andean and Mesoamerica gene pools are considered two big gene pools of common bean and their geographical structure is evident from their domestic forms [Citation16]. The Peru-Ecuador gene pool is considered the third gene pool and contains wild populations first discovered in the 1980s and mainly grown in a very small area of Andes western slopes [Citation21]. The Peru-Ecuador gene pool is mainly characterized by its seed protein named Phaseolin type I, which is absent in the Andean and Mesoamerican gene pool [Citation22].
Domestication is a very complicated process that plays an important role in the modification of wild relatives into a very useful crop [Citation23]. Domesticated forms of common bean can be differentiated from their wild relatives on the basis of different important characteristics such as growth habit, seed dormancy, photoperiod sensitivity, color, shape and size of the plant. These modifications improve the agronomic performance in different climatic conditions and make the crop plant genetically different from the wild forms [Citation24].
Mesoamerica and the Andes were the main regions where domestication of common bean occurred independently. Domestication events occurring in common bean are still under debate by scientists. Some studies are in the favor of a single domestication event [Citation25–27], while some are in favor of two domestication events that have led to establishment of two gene pools [Citation28–30]. The domesticated Mesoamerican gene pool clearly exhibited reduced genetic diversity in different studies [Citation27, Citation31,Citation32]. There was three-fold greater genetic diversity reduction in the Mesoamerican gene pool compared to the Andean gene pool [Citation33]. Very recently Trucchi et al. [Citation34], investigated the temporal dynamics of genetic diversity and selection throughout the domestication process of the common bean in the southern Andes. It was reported that the genomic variation of the pool of cultivars from different Andean regions is higher than the pool of ancient seeds from the central-western and north Argentina.
Diffusion of common bean from its origin to the rest of the world
Several introductions of common bean to different countries were performed with the passage of time. Common bean is now cultivated all over the world and serves as a source of food for millions of people. In Europe, 70% of common bean belongs to the Andean gene pool [Citation35], while the Mesoamerican gene pool is more frequently present in Brazil compared to the Andean gene pool [Citation36]. In China, the Mesoamerican gene pool is more widely spread compared to the Andean gene pool [Citation37], whereas the share of both gene pools is equal in Africa [Citation38]. Additionally, out of the domestication regions, there was less focus on the spatial differentiation between both these gene pools, which resulted in the novel phenotypic and genotypic traits in these gene pools due to hybridization, especially in Europe [Citation35,Citation38].
Common bean genetic resources
Domesticated Phaseolus species (P. vulgaris; P. coccineus; P. dumosus: P. acutifolius and P. lunatus) and several wild Phaseolus species are preserved in different gene banks. Centro Internacional de Agricultura Tropical (CIAT) Cali, Colombia, preserves the world’s largest and most diverse collection of beans with 37,938 Phaseolus accessions belonging to 44 taxa, and it holds 71.6% of the preserved bean germplasm (). Some other stations conserving common bean accessions are Institut für Pflanzengenetik und Kulturpflanzenforschung in Germany (IPK: http://www.ipk-gatersleben.de), the Centro Nacional de Recursos Genéticos e Biotecnologia (CENARGEN/EMBRAPA) in Brazil with more than 1800 common bean accessions (https://www.embrapa.br/en/recursos-geneticos-e-biotecnologia), the National Botanic Garden of Belgium with 2074 accessions from 231 taxa of the Phaseolus tribe (http://www.plantentuinmeise.be/RESEARCH/COLLECTIONS/LIVING/PHASEOLUS/index.html), and Western Regional Plant Introduction Station (WRPIS) in the USA with more than 20,000 Phaseolus accessions - with P. vulgaris as the most abundant species - with 17,000 accessions (https://www.ars.usda.gov/research/publications/publication/?seqNo115=319341). A mini-core collection of nearly 750 Turkish common bean landraces has been recently collected from the different geographical regions of Turkey in different projects funded by The Scientific and Technological Research Council of Turkey. This mini-core collection is maintained at the Abant Izzet Baysal University, Bolu (Turkey) and Sivas University of Science and Technology, Sivas (Turkey), and is available for the bean scientist interested in the Turkish bean germplasm.
Table 1. Genetic resources of various Phaseolus species conserved in various originations all over the world.
A large number of mapping populations have been developed to investigate various traits of interest in common bean [Citation39]. Recombinant inbred (RI) populations from BAT93 x Jalo EEP558 have been taken as core mapping populations because markers resulting from other linkage maps have been mapped in this population [Citation40,Citation41]. Most populations have been produced by making crosses between Mesoamerican and Andean gene pools. For this purpose, diverse parents have been selected exhibiting enough phenotypic variations for agronomic traits, disease resistance, antioxidants, and cooking properties, and also have shown higher polymorphism levels [Citation11, Citation42].
Breeding activities in common bean
Classical breeding
Several studies have been conducted for the successful breeding of common bean using classical breeding methods such as pedigree, single seed descent (SSD), recurrent selection, gamete selection and backcross method to accomplish the wide arrays of objectives. Main breeding efforts have been done to increase the adaptation of the crop to different environmental conditions, to develop genetically improved cultivars and to increase resistance against various biotic and abiotic stresses [Citation43,Citation44]. Common bean is a crop with a low outcrossing proportion [Citation45] and its outcrossing is widely affected by environmental factors [Citation46]. The pedigree method is the most commonly used breeding method for common bean because it facilitates qualitative trait selection in early F2-F4 generations, while this selection would be continued for complex traits like yield up to F5-F7 generation [Citation47]. The single seed or single pod selection is another method that is much faster than the pedigree method and can also be used in the greenhouse easily. Urrea and Singh [Citation48] developed a single seed descent method in common bean by modifying the pedigree method. This method has been widely used by many breeders to develop the RILs (recombinant inbred lines) for various genetic studies in common bean, while no selection is performed in the RILs development used in QTLs studies [Citation49].
Recurrent selection is another important breeding method, and it is mostly used in the breeding of cross-pollinated crops where pollens are present in the ample quantity in the field. However, it is difficult to make good numbers of crosses between two diverse parents in highly self-pollinated crops like beans to produce enough variations for the next cycle of selection [Citation47]. This method has been successfully applied to introduce plant architecture traits into large-seeded decumbent type-III Durango race pinto beans from type-II Mesoamerican black bean race [Citation50]. Sierra pinto variety was the first variety developed via this method with large size seeds of pinto beans and upright type-II short vine habit [Citation51]. Development of Sierra pinto variety served as a genetic bridge for the development of other medium seeded Durango and Jalisco race varieties, and breeders could develop other seed types like Sedona pink bean [Citation52], Merlot small red variety [Citation53] and Matterhorn Great Northern [Citation54].
The selection for multiple traits in a population is possible by crossing the various parents using the gamete selection [Citation55–57]. It was determined to be successful in the development of disease resistant breeding line [Citation58]. The bc-3 allele for bean common mosaic necrosis virus (BCMNV) resistance and bgm gene for bean golden yellow mosaic virus (BGYMV) resistance have been identified through this method [Citation59].
The backcross method is a very useful breeding method mainly used to introduce one or more genes in a variety that lacks such genes [Citation47]. This method has been successfully applied to introduce favorable genetic variations from wild and landraces bean material with introgression between diverse common bean gene pools [Citation60,Citation61]. Intermating between incompatible Phaseolus species has been enhanced by applying the congruity backcross method [Citation62]. The congruity backcross method has been used to produce more recombinant events by crossing common bean with tepary bean and their progenies contain genes from both species [Citation63,Citation64].
Advancement in breeding methodologies for common bean
Advancement in DNA molecular markers and sequencing technologies revolutionized and increased the success of breeding activities when compared to the classical breeding methods [Citation65]. Common bean is a diploid plant (2n = 22) with a genome size of 587 Mb [Citation14] and is beneficial in the genome study of crops with higher genome size like soybean [Citation10]. Common bean genetic map has been available since the 1990s by using different types of molecular markers, i.e. restriction fragment length polymorphism (RFLP) markers [Citation66] and random amplified polymorphic DNA (RAPD) markers [Citation67]. The common bean genome was sequenced by Schmutz et al. [Citation14] through the application of the whole-genome shotgun sequencing technique in common bean genotype “G19833”. The resulting sequences were assembled on 11 chromosomes and ultimately expressed about 80% of the 587-Mb common bean genome. Assembled genome sequences were annotated through the application of ab initio approaches with transcriptome data. A total of 31.632 protein-coding sequences were identified by the 4441 alternately slipped transcripts with 27197 gene models. Moreover, it was reported that transposable elements were present in large amounts and they constituted almost 45% of the genome. The genome of common bean landrace BAT93 was also sequenced by Vlasova et al. [Citation68] under the PhasIbeAm consortium. Based on genetic contents, common bean genome contains some surprises, for instance, it encodes 27,000 genes with a high level of transposons insertion because of the fact that 91% of common bean genes are situated within synteny blocks of soybean. On the other hand, genome duplication is lesser in common bean as compared to the soybean because whole-genome duplication (WGD) occurred in soybean [Citation14]. Recently, the first chromosome-scale version of common bean has been developed by The Joint Genome Institute, Department of Energy) (http://www. phytozome.net/common bean), which is very helpful in common bean genome mapping and marker development. Especially, this database is a very useful database in gene identification studies because it shows the exon and intron regions within the common bean separately.
Common bean genomic resources
Genome sequencing of common bean has helped in understanding the genomics of not only common bean but also soybean [Citation14]. The first linkage map in common bean was developed in the 1990s by using RFLPs and RAPD markers [Citation41, Citation69]. Microsatellite markers are very useful for genetic studies, and GenBank sequences and enriched genomic libraries have been used in the development of simple sequence repeats (SSRs). Yu et al. [Citation70,Citation71] firstly developed the GenBank-based microsatellites generating a set of 38 SSR markers that led to the additional development of 57 SSR by Blair et al. [Citation40] and 20 SSRs by Guerra-Sanz [Citation72]. In common bean, more than 2,000 SSRs markers have been developed from various genomic sequences [Citation40, Citation71, Citation73,Citation74]. Furthermore, size-fractionated bacterial artificial chromosome (BAC) libraries were used by Caixeta et al. [Citation75] for the development of microsatellite markers linked with angular leaf spot resistance gene. BAC libraries with large DNA insertions have been very beneficial in the development of physical maps and can play a vital role in positional cloning, functional analysis, characterization of the genome for gene structure and transgenic studies [Citation76,Citation77]. These libraries have been used in the cloning of large-sized (100–150 kb) genomic fragments as a vector and applied in various economically important species [Citation78].
Genotyping technologies for common bean
The use of genetic approaches for the investigation and analysis of genetic basis having an association with phenotype has greatly facilitated the improvement of the traits of interest. Advancement in genotyping and sequencing technologies revolutionized the breeding activities for various crops [Citation65]. With the emergence of next generation sequencing (NGS) technologies, whole-genome sequencing data and millions of genome-wide single nucleotide polymorphisms (SNPs) for high-throughput genotyping became available for a variety of genetic studies [Citation79]. Various SNP-genotyping platforms like Taqman, SNPlex, BioMark HD, KASPar, Axiom Biobank, Infinium II, GoldenGate, and iPlex have been developed, and successfully used for the genotyping of the various crop [Citation80]. Specific to common bean Blair et al. [Citation81], reported the first 736 SNP chip. Similarly Song et al. [Citation82], developed the BARCBean6K_1 BeadChip with >5000 SNPs, which have been successfully used in various studies Citation83–85]. Moreover, genetic studies in common bean have been also performed using other genotyping platforms like Genotyping-by-sequencing (GBS) [Citation86], Fluidigm platform (www.fluidigm.com) or by KASP genotyping at LGC Genomics service provider (http://www.lgcgroup.com, Citation87].
Development of linkage maps in common bean
Many researchers have conducted studies based on linkage analysis to determine locations of important genes on the related chromosomes of common bean. Freyre et al. [Citation41] first conducted a study to a generate a linkage map for common bean and determined 1258.8 cM genetic distance with 11 linkage groups (LGs). A total of 413 loci are distributed across these 11 LGs having 3.0 cM average distance between neighboring loci. In this study, they developed a RI population (known as BJ population) by crossing BAT93 (Middle American) and Jalo EEP558 (Andean landrace). This BJ population is considered a universal common bean mapping population and has been successfully used to develop linkage maps and other breeding studies in common bean [Citation10, Citation49, Citation88–97]. Yu et al. [Citation71] also developed a common bean linkage map with SSRs using similar populations. Blair et al. [Citation40] constructed a genome-wide anchored microsatellite map from DOR364 x G19833 populations and they identified 150 SSRs. It is known that more than 25 linkage maps have been developed for common bean [Citation10]. These genetic maps play a critical role in the development of various types of markers. On the other hand, advancements in sequencing technologies reduced the genotyping cost and resulted in developing millions of SNP markers for common bean [Citation81, Citation8–101]. The powerful genetic tool has led to the improvement of huge scale molecular markers and the determination of numerous marker–trait associations. Higher numbers of markers resulted in the dense genetic maps that are very helpful for precise localization of major genes and identification of different QTLs controlling various traits of interest [Citation102].
Common bean breeding for biotic and abiotic stresses
More than 200 biotic stress factors causing significant yield and quality losses in common bean have been reported so far [Citation103,Citation104]. Antrachnose (ANT, caused by Colletotrichum lindemuthianum), angular leaf spot (ALS, caused by Phaeoisariopsis griseola), common bacterial blight (CBB), bean golden mosaic virus (BGMV) and bean common mosaic virus (BCMV) are known as the most significant bean diseases worldwide [Citation05–107]. Concerning biotic and abiotic stress factors in common bean, numerous quantitative trait loci (QTL) mapping studies have been performed after development of different molecular techniques. For example, several QTLs about white mold (Sclerotinia sclerotiorum (Lib.) de Bary) as a major constraint to common bean were reported by Park et al. [Citation91] Kolkman and Kelly [Citation94], Ender and Kelly [Citation108], Mkwaila et al. [Citation109] and Soule et al. [Citation110]. These studies have given valuable information for this disease in common bean. Soule et al. [Citation110] identified QTLs (WM4.2, WM5.3, WM5.4 and WM7.3) first time having an association with white mold. Another important biotic factor is angular leaf spot (ALS) caused by the hemibiotrophic fungus Pseudocercospora griseola (Sacc.) Crous and U. Braunis that significantly affects the yield of common bean. Oblessuc et al. [Citation111] identified seven QTLs at ALS effects, four of which were mapped firstly in their research. In another study Keller et al. [Citation112], discovered a region containing 36 candidate genes for ALS resistance. In another realm, two QTLs (FRR2.2CM and FRR2.3CM) located on Pv02 associated with fusarium root rot, which is a soil-borne disease that constrains common bean yield, were reported by Wang et al. [Citation113]. Regarding thrips resistance (Thrips palmi Karny), only a QTL named as Tpr6.1 located on LG b06 was determined by Frei et al. [Citation114]. Concerning the bean weevil (Acanthoscelides obtectus [Say]) which is a crucial postharvest pest of common bean seed Kamfwa et al. [Citation115], discovered three QTLs on chromosomes Pv04 and on Pv06. In addition to these QTLs examples related to biotic stress factors, several QTL studies were also released about abiotic stress factors. For instance Diaz et al. [Citation87], detected 143 QTLs under different stress treatments (drought, P, and Al stresses). Similarly Dramadri et al. [Citation116], identified 18 QTLs, especially those for partitioning and seed yield under drought stress. In another very recent report Sedlar et al. [Citation117], identified QTLs in moderate drought treatment (Wp10.1, Wp1.1, Wp1.2, Wp1.3, Wp5.1, ΦPSII5.1, and ΦPSII9.1) and severe drought treatment (Wp6.2, Wp8.1, Wp9.1, Wp1.2, Wp3.1, Wp7.1, ΦPSII3.1, ΦPSII7.1, and ΦPSII11.1) for leaf water potential (Wp) and effective quantum yield of PSII (ΦPSII). Additionally, extensive QTLs data obtained from previous studies are presented in . Also, extensive data for SCAR markers developed to track many significant diseases of common bean are freely accessible on http://bic.css.msu.edu/_pdf/SCAR_Markers_2010.pdf. Such findings will undoubtedly assist breeders in efforts to improve common bean cultivars resistant to biotic and abiotic stresses with super high yield.
Table 2. Biotic and Abiotic stress related QTLs/markers in common bean.
Common bean breeding for agronomic traits
Genetic resources are fundamental in improving agricultural productivity. These resources contain a variety of alleles necessary for resistance and tolerance to the different diseases, pests and harsh environments found in their natural habitat, and can be used in various breeding programs. One of the main objectives of the common bean breeding programs is to develop high-yielding cultivars with better quality [Citation43]. Previous studies reported that yield-related traits are controlled by several complex genes [Citation148–150]. During the last twenty years, many QTLs about various agronomic traits in common bean have been identified. In addition, few scientific groups [Citation38, Citation151] were interested in the nutrition traits QTLs of common bean worldwide. A glimpse of the diverse applications of QTLs/genes for various agronomic traits in common bean research is presented in . Even though various researchers have determined QTLs with the same chromosomal locations for different agronomic traits, further screening is needed to pinpoint the candidate genes for breeders. Finally, compiling all valuable genomic information related to QTL traits herein will help plant breeders to improve cultivars having higher-yield and resistance to biotic and abiotic factors to meet the diverse demands of humankind.
Table 3. Agronomic traits related QTLs/markers in common bean.
Genome wide association studies (GWAS) in common bean
Dissecting the genetic control of traits of interest is of pivotal importance to foster common bean breeding and to develop new varieties able to adapt to changing climatic conditions. Genome Wide Association Studies (GWAS) is considered the next step, after QTL mapping, to investigate the genetic basis having significant association with traits of interest. During the last decade, a good number of GWAS studies have been conducted in different genetic resources to uncover the genetic basis controlling various traits like flowering, cooking time, seed traits, mineral and antioxidant activity. Cichy et al. [Citation83] performed a study with a similar approach in a panel of 206 P. vulgaris accessions to investigate the genetic diversity and cooking time variations, and focused on chromosomal regions distributed on chromosomes Pv02, Pv03, and Pv06 associated with cooking time. Nemli et al. [Citation164] also reported a study with genotyping by sequencing (GBS) approach in 173 common bean accessions for identification of SNPs associated with various pod traits. They identified 43,018 SNPs markers, and 45 of the SNPs showed significant associations with different pod traits. In addition to these reports Moghaddam et al. [Citation165], performed a study using a panel of 280 modern bean genotypes from race Mesoamerica and investigated ∼30 candidate genes controlling various agronomic traits. Perseguini et al. [Citation166] performed a GWAS for quantitative resistance loci (QRL) controlling resistance to the anthracnose (ANT) and angular leaf spot (ALS). They reported that statistically significantly 21 and 17 associations were determined for ANT and ALS, and the markers SSR-IAC167 and PvM95 located on chromosome Pv03 and the SNP scaffold00021_89379 were associated with both diseases. Ferreira et al. [Citation167] conducted a GBS study for the investigation of introgressed genomic regions associated with the anthracnose, bean common mosaic virus and bean common mosaic necrosis virus. They investigated 12,697 SNPs distributed on different chromosomes and their results verified the positions of resistance genes (I, Co-3, bc-3, and Co-2) on the chromosomes Pv02, Pv04, Pv6, and Pv11, respectively. In addition to these examples, various GWAS conducted to identify genetic loci linked to drought-related traits and drought resistance in common bean [Citation168–171]. Wu et al. [Citation171] identified 196 association loci containing 230 candidate SNPs as being linked to drought resistance in common bean. On the other hand Leitão et al. [Citation172], determined a total of 133 SNPs for transpiration rate, net CO2 assimilation rate, stomatal conductance, chlorophylls a/b, carotenes, and xanthophyll contents. Ninety of these associations were found under water-deficit conditions. To the best of our knowledge, the most notable GWAS studies conducted in common bean are provided in . To sum up, important breakthroughs in the field of GWAS in recent years have taken common bean research to a new dimension. These resources will provide a new framework for breeders to accelerate the common bean improvement.
Table 4. Genome-wide association studies for common bean to investigate the genetic basis associated with trait of interests.
Kompetitive allele specific PCR (KASP) technology in common bean
Kompetitive Allele Specific PCR (KASP) is a modern technique of SNP genotyping requiring only a few SNP markers to genotype various samples. This technology was developed by LGC Genomics Ltd., based on fluorescent signals, and emerged as an efficient and very low-cost genotyping method [Citation198,Citation199]. In common bean, very few numbers of KASP genotyping studies have been conducted. For example Hurtado-Gonzales et al. [Citation200], aimed to develop highly specific, tightly linked, effective molecular markers having an association with Ur-3 rust resistance gene in common bean. They found that SS68 KASP marker has an association with Ur-3 rust resistance gene. They performed the validation of this marker on a panel of 130 diverse common bean, evaluated SS68 KASP as a highly accurate marker having no false results. Bean yellow mosaic virus (BYMV) is a major limitation to common bean production. Hart and Griffiths [Citation86] aimed to develop markers having resistance to this important disease. They found a total of 44 SNP markers having an association to the phenotype and they converted seven of these markers into KASP assay and found tight linkage of these markers to BYMV resistance in an F2 population of 185 individuals. Anthracnose is considered one of the most destructive diseases of common bean. Recently Gilio et al. [Citation201], aimed to perform fine mapping for anthracnose-resistance locus and to develop markers tightly linked to this locus. They found KASP markers ss56 and ss92 the as most tightly linked markers to Co-AC locus.
Functional genomics studies in common bean
Functional genomics (FG) is crucial for crop improvement and provides a deep insight into molecular plant breeding by classifying the expressed genes, metabolites and proteins related to specific characteristics [Citation202]. FG helps to identify the genes regulating crop improvement, resistance to abiotic/biotic stress factors, yield and multiple different components influencing the economic importance in legumes. A cell contains a complete set of transcriptomes, so understanding the transcriptome is very beneficial for interpreting the functional elements of the genome. Transcriptomics is mainly applied to investigate the transcriptional structure of genes, changes in each transcript due to various conditions. Different types of techniques have been developed and applied in various crops to deduce and quantify the transcriptome [Citation203]. As common bean is an important legume crop, a lesser amount of short expressed sequence tags (ESTs) was only identified with conventional processes. Hatey et al. [Citation204] described ESTs as partial sequences of transcribed genes that represent gene expression in various tissues in which mRNA was obtained in different genotypes. Various studies have been conducted to create cDNA libraries for various legume crops [Citation205–208] and stated that the number of ESTs currently available for all plant species are more than 21 million sequences. More than 3 million of all sequences have been produced from legumes. Development of ESTs to common bean started with moderate amounts of GenBank entries by groups from CIAT-Colombia, UNESP-Brazil and UNAM-Mexico organizations [Citation209–211]. Ramirez et al. [Citation211] served as the starting point for the common bean functional genomics. The project conducted by Ramirez et al. [Citation211] was basically started to generate EST profiles of P-deficient roots and N2-fixing root nodules. However, EST resources for pods and leaves of common bean was also determined during the project. A total of 21,026 ESTs obtained from five different cDNA libraries containing phosphorus-deficient roots, nitrogen-fixing root nodules, developing leaves and pods of the Negro Jamapa 81 (Mesoamerican genotype). The details of statistics of common bean ESTs are presented in . These sequences were separated into four main sections, such as plant development and cell cycle, metabolism, interaction with the environment, and unknown function. The investigated resources have contributed to genetic and genomic studies in common bean worldwide because these sequences have played a greater role in the understanding of common bean improvement, metabolism, and adaptations to various stresses [Citation33].
Table 5. Sequencing and contigging statistics of common bean ESTs (adapted from Blair et al. [Citation214]).
Melotto et al. [Citation210] used the anthracnose resistant breeding line SEL 1308 as the source of mRNA. cDNA libraries (PVEPLE1, PVEPSE2, and PVEPSE3) were generated from leaves, shoots and inoculated shoots of bean seedlings. A total of 5255 single-pass sequences were involved in the database after selection based on the quality of the sequences. Then, these EST sequences were trimmed and constructed a UniGene collection including 3126 sequences. Among these, 314 unigenes revealed important resemblances to genomic sequences of common bean and ESTs, which shows that 2818 unigenes of the database represent recently identified common bean genes. Additionally, 387 out of all unigenes were determined as common bean specific. Tian et al. [Citation212] constructed the cDNA library to classify genes related to phosphorous starvation where a comparatively well-adapted cultivar to low Pi conditions and Pi-efficient genotype (G19833) was used as material. They identified Pi starvation-responsive genes (+240 putative) and identified clones were sequenced, and BLASTx/BLASTn analysis showed an array of 82 genes revealing a high ratio of sequence homology to known and unknown proteins in the database. Following this, differentially expressed genes were divided into five categories: signaling-transcription, transporter-channel, stress-defense, carbon metabolism and another metabolism.
McClean et al. [Citation213] utilized all available EST sequences to improve contig sequences representing gene space in the genome. Their results revealed that most genes had only one copy in common bean, while there were duplicated genes in the soybean genome. This situation is an indication of the diploid history of common bean compared to the soybean polyploidy background. Similarly, McConnell et al. [Citation10] used new sequence-based sources to characterize SNPs and InDel density in the genome. Sequence data were obtained from 550 gene fragments of two commonly used common bean genotypes (BAT93 and Jalo EEP558) for research activities. They identified over 1,800 SNPs and InDels, 300 of which were screened in the RI population, and 395 polymorphic gene fragments were obtained from nearly 593 kb sequence data. The sequences of BAT93 and Jalo EEP558 were compared with each other, and with the contig sequences to determine SNPs and InDel polymorphisms. Blair et al. [Citation214] conducted a study using full-length cDNA technology to develop ESTs. They constructed the library to relate genes expressed in various conditions, such as low soil phosphorus, drought and high soil aluminum toxicity, using BAT477 and G19833 genotypes. It was determined that 4,219 unigenes were recognized consisting of 1,238 singletons and 2,981 contigs. Nearly half of the sequences were found as unique or represented the 5′ ends of known genes compared to other EST sequencing in common bean . These results can be beneficial to functional gene explanation, investigation of splice site variants, discovery and validation of drought or abiotic stress-associated genes in common bean.
Table 6. Comparison of major EST sequencing efforts in common bean (adapted from Blair et al. [Citation214]).
The ESTs in databases are suitable resources for the identification of EST-derived SSRs. As a good example, 302 new EST-SSR markers from 9.583 ESTs revealing good amplification quality were developed based on transferability and polymorphism information by Garcia et al. [Citation73]. Their results showed that 82% of them were transferable across at least one species. The average PIC value was determined as 0.53-genomic SSRs, 0.47-EST-SSRs, and the average amount of alleles per locus was 4 and 3, respectively. Another set of genomic data was constructed by Kalavacharla et al. [Citation216] as they sequenced many cDNA libraries from different plant tissues (leaves, flowers, pods, and roots) using the Roche 454-FLX pyrosequencing platform. The 59,295 unigenes containing 39,572-contigs and 19,723-singletons were identified, and they provided a substantial transcriptome dataset to common bean. It was determined that 31,664 unigenes had no matches to GenBank and could be thought of as new common bean transcripts. Literally, the study resulted in a 150% increase in the amount of common bean ESTs.
In summary, the EST sequences of common bean present the foundation for genome-wide transcript studies. They are sources of established molecular markers to map linkage groups and anchored to physical maps. Additionally, full-length cDNA technology can be very functional for sequencing of the transcriptome, gene annotation, comparative genomics and identification of the genetic basis of agronomically important traits.
Transcription factors in common bean
Transcription factors (TFs) are important genes synchronizing signal transduction and expression of genes during biotic and abiotic stress responses [Citation176]. Due to the importance of TFs, in the regulation of stress-related genes, investigations on plant TFs have been rising rapidly in recent years. In line with this purpose, various studies have been conducted to identify different TFs and to determine the expression levels of these genes under biotic and abiotic stress conditions in common bean. For instance, Apetala2-ethylene-responsive element binding factor (AP2-ERF) gene family was identified by Kavas et al. [Citation217]. It was noted that the expression levels of 9 PvAP2-ERFs genes were determined in salt-stressed leaf/root tissues. Similarly Kavas et al. [Citation218], classified 155 bHLH (basic helix-loop-helix) genes by using bioinformatics tools and investigated the expression levels of 16 PvbHLH genes to salt stress in the root/leaf tissues. In another study Büyük et al. [Citation219], investigated a total of 24 candidate HSP70 (heat shock protein 70) genes in leaf/root tissues. In a more comprehensive study, a total of 86 NAC (NAM, ATAF1/2 and CUC2) genes were identified by Wu et al. [Citation220]. It was noted that the expression patterns of the 22 NAC genes were varied under drought stress conditions.
Another important TF gene named C2C2-YABBY was identified İnal et al. [Citation221]. They aimed to investigate some potential genes associated with salt tolerance and used two common bean genotypes (sensitive and tolerant) under salt stress conditions and studied the gene expression levels of C2C2-YABBY genes. Concerning the WRKY gene family, 88 WRKY genes were reported, and the response of 19 WRKY genes was discovered under drought stress [Citation177].
SBP (SQUAMOSA promoter binding protein family) genes as other important TF genes were identified by Ilhan [Citation222]. Some of the Phvul-SBP genes in the roots, leaf, and floral organs were up- or down-regulated. Buyuk et al. [Citation223] reported 42 candidate PvDOF genes, and the expression levels of 9 out of the PvDOFs genes were up- or down-regulated under salt stress in different tissues. They also identified CAMTA (The calmodulin-binding transcriptional activator) gene family and investigated the expression levels of genes to figure out some potential genes that confer resistance to salt stress. On the other hand, Whirly and ArrB gene families were studied by Gökdemir [Citation224]. The expression levels of the selected genes (Phv-Why-1, −2, −3 and Phv-ArrB-5, −8, −13, −16, −19) were determined under artificial epidemic of Sclerotinia sclerotiorum in two registered varieties (Önceler and Akman). It was seen that the expression levels of Phv-Why-1, −2, −3 and Phv-ArrB-13, −19 genes were up-regulated in Önceler compared to Akman. On the other hand, Phv-Arr-5, −8, −16 genes were up-regulated in Akman compared to Önceler. However, the change in the expression levels in the Phv-Why-2 gene in both varieties was determined as statistically significant compared to the rest of the genes under biotic stress Zhang et al. [Citation225]. studied the expression levels of six PvB3 genes under salt stress in cotyledon, hypocotyl, radicle, and some B3 family members revealed a relatively high expression in the radicle and hypocotyl.
Another study by Silva et al. [Citation226] investigated the expression levels of genes in genotypes of common bean IAC Imperador (P-responsive) and DOR 364 (P-unresponsive) under different P concentrations. P-responsive genotype reported 1538 up-regulated genes under P restriction and 1679 up-regulated genes in the control level, while the P-unresponsive genotype reported 13 up-regulated genes in the control level and only 2 up-regulated genes under P restriction. Similarly, Hernández et al. [Citation227] reported identifying 372 bean transcription factor (TF) genes. A total of 126 genes showed significant differential expression (response to P: 62%) in P-deficient roots. The studies described above related to transcription will provide significant information for plant breeders and geneticists to conduct genomic studies of the common bean in the near future.
Common bean proteomics
During 1970s and 1980s, most breeding activities were concerned to improve the protein quality in crops by improving the seed protein contents and balancing the composition of different essential amino acids [Citation228]. However, protein quality was less focused on common bean and it contains lesser concentrations of methionine, cysteine, and sulfur amino acid [Citation229]. Phasolin or 7S globulin phaseolin is the most plentiful seed protein and constitutes up to 50% of total protein in common bean [Citation230]. Lectins are the second most abundant protein (5-10% of total protein) and 11S globulin legume present in very low levels in common bean [Citation231]. Several efforts have been done to improve the protein quality in common bean; however, these efforts are dependent on the availability of alleles conferring phaseolin and erythroagglutinating phytohemagglutinin deficiency [Citation232–234]. Bollini et al. [Citation235] identified a unique genetic source for the deficiency of phytohemagglutinin. A very low concentration of sulfur amino acids is present in 7S globulin, and phaseolin levels are positively correlated with the methionine [Citation233]. To improve and balance the seed protein contents, it is necessary to remove the phytohemagglutinin that enhances the phaseoline concentration in the seeds [Citation234].
During the last decade, most common bean studies have been conducted for the improvement of biotic stress, abiotic stress, diversity and seed storage protein. For the separation of protein and its quantification in various plants, 2-dimensional (2-D) gel electrophoresis has been used, followed by either liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) or matrix-assisted laser desorption/ionization-mass spectrometry coupled to time of flight (MALDI-TOF) [Citation236]. Robison et al. [Citation237] identified 640 protein species using two common bean lines, A195 and Sacramento. During abiotic stress, the effects of gaseous pollutant ozone were observed and showed distinct changes in protein in the affected common bean leaves by two-dimensional electrophoresis (2- DGE; [Citation238]. Zadražnik et al. [Citation239] performed various proteomic analyses to investigate the drought stress in the common bean leaves using two cultivars Tiber (less drought sensitive) and Starozagorski čern (more drought sensitive). They identified 58 proteins in Tiber and 64 in Starozagorski čern by using LC-MS/MS analysis. Parreira et al. [Citation240] applied high-throughput gel-free proteomics approach (LC–MS/MS) for the investigation of hallmarks of seed development. They identified 418 proteins and 255 of them were characterized. According to Badowiec and Weidner [Citation241], the response of common bean toward the chilling stress was totally dependent on the exposure of common bean to low temperature and duration of low temperature. They identified various proteomic variations in the root during different periods of low temperature. Salavati et al. [Citation42] used two-dimensional polyacrylamide gel electrophoresis (2-DE) coupled with mass spectrometry (MS) for the proteome analysis of common bean to investigate the symbiosis between common bean roots and bacteria. They identified 483 different proteins, among these only 29 plant and 3 bacterial proteins were associated with early-stage symbiosis. Among the 29 plant proteins, while 19 proteins showed up-regulated expression patterns, 10 showed down regulated expression patterns. These up-regulated proteins were associated with the storage, energy production and protein synthesis; unlike these, down regulated proteins were associated with metabolism. Rust is an important disease in common bean and when rust infected leaves were studied, they showed that R-gene based defense modulates a protein that was identical to the basal defense system [Citation243]. Phenol is an old protein extraction method and now protein extraction is performed with TCA–acetone as a new method and followed by a clean-up step that provides a higher amount of storage and defense protein [Citation236]. Natarajan et al. [Citation244] used the TCA-acetone method with 2-DGE analysis for the analysis of improved common bean by maintaining various changes in protein components. Phaseolin is the major seed storage protein of common bean and phosphorylation is post-translational protein modification (PTM) playing a vital role in proteome complexity López-Pedrouso et al. [Citation245]. performed proteome analysis in two common bean cultivars Sanilac and Tendergreen to investigate the phosphorylation presence and its degradation in the dormant and germinating seeds. They identified remarkable variations in the levels of phosphorylation of the phaseolin from dormancy to seed germination. Mensack et al. [Citation246] performed 2-DGE analysis for the investigation of differences between wild and domesticated common beans and they identified various protein changes between wild and domesticated cultivars. The Database LegProt (http://bioinfo.noble.org/manuscript-support/legumedb) contains the sequences of all legumes, including common bean, and would play a beneficial role in the identification of legumes proteins in a better way [Citation247].
Bioinformatics tools and online functional database resources
Various integrative bioinformatics tools and online databases have been constructed to assemble common bean information and the genomic data related to common bean, such as Phytozome database v12.1 (https://phytozome.jgi.doe.gov/pz/portal.html), Legume Information System (LIS: http://phavu.comparative-legumes.org/gb2/gbrowse/Pv1.0/), KnowPulse (https://knowpulse.usask.ca/), National Center for Biotechnology Information (NCBI-https://www.ncbi.nlm.nih.gov/), Hidden Markov model (HMM, http://www.ebi.ac.uk), Pfam databases (http://pfam.xfam.org/) [Citation248], The decrease redundancy tool (http://web.expasy.org/decrease_redundancy), ProtParam Tool (http://web.expasy.org/protparam), Gene Structure Display Server v2.0 (GSDS) (http://gsds.cbi.pku.edu.cn/) [Citation249], the Multiple EM for Motif Elicition tool (MEME v4.11.1; http://meme-suite.org/) [Citation250], The PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/), the WoLF PSORT (http://www.genscript.com/psort/wolf_psort.html) [Citation251], TargetP 1.1 (http://www.cbs.dtu.dk/services/TargetP/) [Citation252], psRNA Target Server (http://plantgrn.noble.org/psRNATarget/) [Citation253], Blast2GO (http://www.blast2go.com) Software [Citation254], Plant Genome Duplication Database (PGDD; https://chibba.agtec.uga.edu/duplication/) [Citation255], iTAK - Plant Transcription factor & Protein Kinase Identifier and Classifier (http://itak.feilab.net/cgi-bin/itak/index.cgi) [Citation256], Phyre2 database (Protein Homology/Analogy Recognition Engine; http://www.sbg.bio.ic.ac.uk/phyre2) [Citation257] and PAL2NAL (http://www.bork.embl.de/pal2nal) [Citation258]. The above listed online databases have been commonly used in genome-wide studies in common bean. Moreover, a web-accessible TFs database for common bean known as PvTFDB has been developed Gautam et al. [Citation259] with 2370 putative TF gene models distributed in 49 TF families. The tools and databases can provide valuable information to breeders related to gene expression, genome functional components, functional genes, comparative genomics, gene family, genetic maps studies etc. in common bean.
Transgenics/genetic transformation in common bean
Common bean is not a much responsive species to genetic transformation with the objective to investigate the genes and their functions. This is the reason why very few efforts have been done in the transformation of common bean as compared to its genomic mapping, transcriptomic and candidate genes identification [Citation260]. Transformation in common bean has been achieved by direct and indirect methods. Electroporation or particle bombardment is a direct gene transfer method and Agrobacterium tumefaciens is an indirect method [Citation261–265]. According to Aragão et al. [Citation266], exotic genes can be effectively transferred to superficial cell layers when bombardment is performed on the meristematic cells of embryonic axes. An electrical particle acceleration device was used by Russell et al. [Citation267] for the production of transgenic navy bean plants. The protocol of [Citation267] showed limitation as it was very time-consuming with less transformation frequency (0.03%). However, studies by Kim and Minamikawa [Citation264] resulted in the recovery of transgenic bean plants when the bombardment was performed on apical meristematic regions [Citation268,Citation269]. According to the literature, the first gene transformation in common bean was achieved in 1993, and till now very few genes have been introduced for agronomic traits [Citation39]. The be2s1 gene was introduced in common bean to improve the seeds methionine contents. Aragão et al. [Citation270] used five transgenic lines and found that two lines resulted in 14% and 23% increase in methionine content as compare to controlled lines. The bar gene is responsible for the coding of phosphinothricin acetyl transferase (PAT) and it confers resistance against the herbicide glufosinate ammonium and phosphinothricin. The introduction of the bar gene was performed by Russell et al. [Citation267] and for the production of virus-resistant plants; they also introduced the coat protein gene from bean golden mosaic geminivirus (BGMV). While Faria et al. [Citation271] used mutated AC1 viral gene for the production of plants having higher resistance against the BGMV and Bonfim et al. [Citation272] used RNA interference (RNAi) to produce higher resistance against the BGMV. Molecular characterization of the first commercial transgenic common bean was performed by Aragão et al. [Citation273]. According to Aragão et al. [Citation273], this transgenic common bean showed immunity to BGMV and when crossed with a non-transgenic commercial variety, it exhibited stability of transgenes up to eight self-pollinated generations. Rep-TrAP-REn and BC1 genes were transferred in the common bean to produce resistance against golden mosaic geminivirus (BGMV-BR) by Aragão et al. [Citation274] and transgenic lines showed resistance against this disease.
Agrobacterium-mediated transformation remained unsuccessful as compared to particle bombardment in common bean [Citation275,Citation276] because it was recalcitrant to Agrobacterium due to poor regeneration in tissue culture [Citation277–278]. Genga et al. [Citation279] used different strains of Agrobacterium in the transformation of cotyledonary node and primary leaf explants, and they produced callus on kanamycin selection media, but they failed to obtain full explants. A. rhizogenes strain A4RS and A. tumefaciens strain C58Z707/pGA482 were applied by McClean et al. [Citation280] for the transformation of cotyledons and hypocotyls; however they failed to regenerate the full plant successfully. Franklin et al. [Citation275] used the A. tumefaciens strain EHA 101 for transformation in kidney bean and they produced GUS positive callus from it. Lewis and Bliss [Citation281] used C58 strain with stab inoculation for the transformation of various shoot types of meristematic regions; however they were unsuccessful in regenerating any shoots during their study. Common bean intact leaves were transferred by Kapila et al. [Citation282] using the vacuum infiltration of Agrobacterium for transient gene expression studies and they found higher GUS expression (20-90%). Embryo axis explants were applied by Mukeshimana et al. [Citation283] for the common bean transformation and obtained chimeric plants, which were unsuccessful to acclimatize in the soil. According to Estrada-Navarrete et al. [Citation284] A. rhizogenes resulted in a higher transformation level (75-90%) as compared to Agrobacterium tumefaciens. Estrada-Navarrete et al. [Citation284] developed an easy and efficient protocol of gene transformation for common bean. Several common bean accessions, landraces and cultivars were used in their study, which resulted in 75-90% transformation efficiency. Four strains of A. rhizogenes were used in their study and root hairs were effectively induced due to strain K599. Later in 2007, a detailed and more effective protocol for gene transformation in common bean with strain K599 was presented by Estrada-Navarrete et al. [Citation285]. Colpaert et al. [Citation278] developed a composite method for the production of common bean having transgenic roots. In a very recent study reported by Xue et al. [Citation286], they identified a PvPOX1 gene from CAAS260205 (Fusarium wilt resistant genotype) and transferred the resistant allele in to BRB130 (Fusarium wilt susceptible genotype) through the root hairs with the help of Agrobacterium rhizogenes. Liu et al. [Citation287] developed a transformation protocol by combining sonication and vacuum infiltration methods using A. tumefaciens. This approach is also known as Sonication assisted Agrobacterium-mediated Transformation (SAAT) and it resulted in 12% transformation in common bean. The study by [Citation288] resulted in the higher transformation efficiencies between 10-28% with the A. tumefaciens.
Several protocols have been developed for the regeneration of common bean from the meristem cells based on the direct organogenesis. Intact seedling and cotyledonary nodes were used from two common bean cultivars Fonix and Maxidor to regenerate full plants [Citation289,Citation290]. In other studies, embryonic axis explants were used to regenerate full plants [Citation291–292]. Veltcheva and Svetleva [Citation293] selected three Bulgarian common bean varieties and used their leaf petioles for the plant regeneration and found genotype dependent reactions. The regeneration process in 10 common bean cultivars using apical meristems was studied by Sabzikar et al. [Citation294] and they found that the race of the cultivar has a direct effect on the multiplication of apical shoot meristem. Apical meristem and cotyledonary node explants were used by Arellano et al. [Citation277] for the regeneration of 10 different common bean cultivars using indirect organogenesis, but they found a low regeneration frequency in their study.
Current hot topics in common bean and future directions
A good number of studies QTL/linked markers regarding agronomic, cooking quality, biotic and abiotic stresses are available. There is need to validate these identified genetic bases for their usage in marker assisted breeding of common bean.
In wheat 660K SNP array, in potato 12K, 20K and 22K SNP chips are available. These arrays techniques fasten the breeding activities in these crops. As a nutritionally potential crop, there is a need to develop such type of arrays having a higher number of SNPs for speed breeding in common bean.
Kompetitive Allele Specific Polymerase Chain Reaction (KASP) assays have been used by some scientists in common bean. There is a need to develop and validate the KASP assay for genes that underpin economically important traits in common bean including adaptability, grain yield, quality and biotic and abiotic stress resistance.
Characterization of genetic resources is considered as a prerequisite of breeding activities. There is a need to collect, conserve and characterize the genetic resources to explore novel variations for future breeding.
Common bean can be crossed with its relative species like tepary bean and also confirmed the occurrence of useful genes that can be used for the common bean improvement [Citation295]. Therefore, there is a need to cross the common bean with its wild relatives to increase the chance of incorporating useful novel chromosomal regions for accelerated breeding.
There is a scarcity of information regarding the interaction among various stresses. There is a need to conduct studies in common bean to investigate the combined effects of various stresses like salt versus drought, heat versus drought, drought × salt × nutrition, among others.
CRISPR/Cas has opened a new window for plant breeders in recent years. However, to our knowledge, till now CRISPR/Cas has not been used in common bean by the scientific community. This technique can be used to generate a broad range of genetic diversity for common bean breeding in an unprecedented way and can be utilized to develop modern cultivars resistant to biotic and abiotic stress factors by plant breeders.
Conclusions
Common bean, known as “poor man’s meat”, is a globally important legume crop and appeals both to farmers and consumers. As the world is confronting with simultanious problems of climate change and rapidly increasing population, there is a need to utilize various breeding and biotechnological tools for the development of climate resilient cultivars. Regarding these factors, present efforts comprehensively reported limiting factors, previous and ongoing efforts for the study of common bean in the field of structural and functional genomics, transcriptomics, gene transformation, genome editing and proteomics. We envisage that the information presented herein will be helpful for the breeding community to take more efforts to serve the world population with high quality food in sufficient quantity [Citation153, Citation296,Citation297].
References
- Alexandratos N, Bruinsma J. World agriculture towards 2030/2050: the 2012 revision. 2012.
- Beebe SE, Rao IM, Mukankusi CM, et al. Improving resource use efficiency and reducing risk of common bean production in Africa, Latin America, and the Caribbean. Centro Internacional de Agricultura Tropical (CIAT). 2012.
- Akibode CS, Maredia MK. Global and regional trends in production, trade and consumption of food legume crops (No. 1099-2016-89132). 2011.
- Yeken MZ, Kantar F, Çancı H, et al. Breeding of dry bean cultivars using Phaseolus vulgaris landraces in Turkey. Int J Agric Wildl Sci. 2018;4(1):45–54.
- Aydin MF, Baloch FS. Exploring the genetic diversity and population structure of Turkish common bean germplasm by the iPBS-retrotransposons markers. Legum Res. 2019;42(1):18–24.
- Nadeem MA, Habyarimana E, Çiftçi V, et al. Characterization of genetic diversity in Turkish common bean gene pool using phenotypic and whole-genome DArTseq-generated silicoDArT marker information. PLoS ONE. 2018;13(10):e0205363.
- Nadeem MA, Gündoğdu M, Ercişli S, et al. Uncovering phenotypic diversity and DArTseq marker loci associated with antioxidant activity in common bean. Genes. 2020;11(1):36.
- Nadeem MA, Karaköy T, Yeken MZ, et al. Phenotypic characterization of 183 Turkish common bean accessions for agronomic, trading, and consumer-preferred plant characteristics for breeding purposes. Agronomy. 2020;10(2):272.
- Nadeem MA, Çilesiz Y, Ali F, et al. Investigation of quality and cooking traits diversity in a global common bean germplasm. Glob J Bot Sci. 2020;8(1):21–29.
- McConnell M, Mamidi S, Lee R, et al. Syntenic relationships among legumes revealed using a gene-based genetic linkage map of common bean. Theor Appl Genet. 2010;121(6):1103–1116.
- Broughton WJ, Hernandez G, Blair M, et al. Beans (Phaseolus spp.)–model food legumes. Plant Soil. 2003;252(1):55–128.
- Freytag GF, Debouck DG. Taxonomy, Distribution, and Ecology of the Genus Phaseolus (Leguminosae-Papilionodeae) in North America, Mexico and Central America. Taxonomía, distribución y ecología del género Phaseolus (Leguminosae-Papilionodeae) en Norteamérica, México y Centroaméric. SIDA, Botanical Miscellany. 2002.
- WHO. [cited on 2021 Feb 14]. 2020.
- Schmutz J, McClean PE, Mamidi S, et al. A reference genome for common bean and genome-wide analysis of dual domestications. Nat Genet. 2014;46(7):707–713.
- Stefanović S, Pfeil BE, Palmer JD, et al. Relationships among phaseoloid legumes based on sequences from eight chloroplast regions. Syst Bot. 2009;34(1):115–128.
- Bellucci E, Bitocchi E, Rau D, et al. Genomics of origin, domestication and evolution of Phaseolus vulgaris. In: Tuberosa R, Graner A, Frison E, editors. Genomics of plant genetic resources. Dordrecht: Springer; 2014. p. 483–507.
- Lavin M, Herendeen PS, Wojciechowski MF. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst Biol. 2005;54(4):575–594.
- Galeano CH, Fernández AC, Gómez M, et al. Single strand conformation polymorphism based SNP and Indel markers for genetic mapping and synteny analysis of common bean (Phaseolus vulgaris L.). BMC Genom. 2009;10(1):614–629.
- Delgado-Salinas A, Bibler R, Lavin M. Phylogeny of the genus Phaseolus (Leguminosae): a recent diversification in an ancient landscape. Syst Bot 2006;31(4):779–791.
- Bitocchi E, Nanni L, Bellucci E, et al. Mesoamerican origin of the common bean (Phaseolus vulgaris L.) is revealed by sequence data. Proc Natl Acad Sci USA. 2012;109(14):E788–E796.
- Debouck DG, Toro O, Paredes OM, et al. Genetic diversity and ecological distribution of Phaseolus vulgaris (Fabaceae) in northwestern South America. Econ Bot. 1993;47(4):408–423.
- Kami J, Velásquez VB, Debouck DG, et al. Identification of presumed ancestral DNA sequences of phaseolin in Phaseolus vulgaris. Proc Natl Acad Sci USA. 1995;92(4):1101–1104.
- Di Vittori V, Bellucci E, Bitocchi E, et al. Domestication and crop history. In de la Vega MP, Santalla M, Marsolais F, editors. The common bean genome. Cham: Springer; 2017. p. 21–55.
- Gepts P, Papa R. Evolution during domestication. 2003. 10.1038/npg.els.0003071
- Kwak M, Gepts P. Structure of genetic diversity in the two major gene pools of common bean (Phaseolus vulgaris L., Fabaceae). Theor Appl Genet. 2009;118(5):979–992.
- Papa R, Gepts P. Asymmetry of gene flow and differential geographical structure of molecular diversity in wild and domesticated common bean (Phaseolus vulgaris L.) from Mesoamerica. Theor Appl Genet. 2003;106(2):239–250.
- Rossi M, Bitocchi E, Bellucci E, et al. Linkage disequilibrium and population structure in wild and domesticated populations of Phaseolus vulgaris L. Evol Appl. 2009;2(4):504–522.
- Acosta-Gallegos JA, Kelly JD, Gepts P. Prebreeding in common bean and use of genetic diversity from wild germplasm. Crop Sci 2007;47:S-44.
- Angioi SA, Desiderio F, Rau D, et al. Development and use of chloroplast microsatellites in Phaseolus spp. and other legumes. Plant Biol (Stuttg)). 2009;11(4):598–612.
- Papa R, Nanni L, Sicard D, et al. The evolution of genetic diversity in Phaseolus vulgaris L. New approaches to the origins, evolution and conservation of crops. In: Motley TJ, Zerega N, and Cross H, editors. Darwin’s harvest. New York (NY): Columbia University Press; 2006. p. 121–142.
- Bitocchi E, Bellucci E, Giardini A, et al. Molecular analysis of the parallel domestication of the common bean (Phaseolus vulgaris) in Mesoamerica and the Andes. New Phytol. 2013;197(1):300–313.
- Nanni L, Bitocchi E, Bellucci E, et al. Nucleotide diversity of a genomic sequence similar to SHATTERPROOF (PvSHP1) in domesticated and wild common bean (Phaseolus vulgaris L.). Theor Appl Genet. 2011;123(8):1341–1357.
- Bellucci E, Bitocchi E, Ferrarini A, et al. Decreased nucleotide and expression diversity and modified coexpression patterns characterize domestication in the common bean. Plant Cell. 2014;26(5):1901–1912.
- Trucchi E, Benazzo A, Lari M, et al. Ancient genomes reveal early Andean farmers selected common beans while preserving diversity. Nat Plants. 20217:1–6.
- Angioi SA, Rau D, Attene G, et al. Beans in Europe: origin and structure of the European landraces of Phaseolus vulgaris L. Theor Appl Genet. 2010;121(5):829–843.
- Burle ML, Fonseca JR, Kami JA, et al. Microsatellite diversity and genetic structure among common bean (Phaseolus vulgaris L.) landraces in Brazil, a secondary center of diversity. Theor Appl Genet. 2010;121(5):801–813.
- Zhang X, Blair MW, Wang S. Genetic diversity of Chinese common bean (Phaseolus vulgaris L.) landraces assessed with simple sequence repeat markers. Theor Appl Genet. 2008;117(4):629–640.
- Blair MW, González LF, Kimani PM, et al. Genetic diversity, inter-gene pool introgression and nutritional quality of common beans. Theor Appl Genet. 2010;121(2):237–248.
- Gepts P, Aragão FJ, De Barros E, et al. Genomics of Phaseolus beans, a major source of dietary protein and micronutrients in the tropics. In: Moore PH, Mong R, editors. Genomics of Tropical Crop Plants. New York (NY): Springer; 2008. p. 113–143
- Blair MW, Pedraza F, Buendia HF, et al. Development of a genome-wide anchored microsatellite map for common bean (Phaseolus vulgaris L.). Theor Appl Genet. 2003;107(8):1362–1374.
- Freyre R, Skroch PW, Geffroy V, et al. Towards an integrated linkage map of common bean. 4. Development of a core linkage map and alignment of RFLP maps. Theor Appl Genet. 1998;97(5-6):847–856.
- Nodari RO, Koinange EMK, Kelly JD, et al. Towards an integrated linkage map of common bean : 1. Development of genomic DNA probes and levels of restriction fragment length polymorphism. Theor Appl Genet. 1992;84(1–2):186–192.
- Beaver JS, Osorno JM. Achievements and limitations of contemporary common bean breeding using conventional and molecular approaches. Euphytica. 2009;168(2):145–175.
- Şen K, Koca AS, Kacar G. Importance, Biology, Damage and Management of Bean Weevil Acanthoscelides obtectus Say (Coleoptera: Chrysomelidae). J Inst Sci Technol. 2020;10(3):1518–1527.
- Brunner BR, Beaver JS. Estimation of outcrossing of the common bean in Puerto Rico. HortScience. 1989;24(4):669–671.
- Ibarra-Perez FJ, Ehdaie B, Waines JG. Estimation of outcrossing rate in common bean. Crop Sci. 1997;37(1):60–65.
- Kelly JD, Cichy KA. Dry bean breeding and production technologies. In: Siddiq M, Uebersax MA, editors. Dry beans and pulses: production, processing, and nutrition. Chichester (UK): Wiley-Blackwell; 2013. p. 23–54.
- Urrea CA, Singh SP. Comparison of mass, F2-derived family, and single-seed-descent selection methods in an interracial population of common bean. Can J Plant Sci. 1994;74(3):461–464.
- Kelly JD, Vallejo VA. A comprehensive review of the major genes conditioning resistance to anthracnose in common bean. HortScience. 2004;39(6):1196–1207.
- Kelly JD, Adams MW. Phenotypic recurrent selection in ideotype breeding of pinto beans. Euphytica. 1987;36(1):69–80.
- Kelly JD, Adams MW, Saettler AW, et al. Registration of ‘Sierra’pinto bean. Crop Sci. 1990;30(3):745–746.
- Kelly JD, Varner GV, Hosfield GL, et al. Registration of ‘Sedona’pink bean. Crop Sci. 2006;46(6):2707–2708.
- Hosfield GL, Varner GV, Uebersax MA, et al. Registration of’Merlot’small red bean. Crop Sci. 2004;44(1):351–353.
- Kelly JD, Hosfield GL, Varner GV, et al. Registration of ‘Matterhorn’great northern bean. Crop Sci. 1999;39(2):589–590.
- Singh SP. Gamete selection for simultaneous improvement of multiple traits in common bean. Crop Sci. 1994;34(2):352–355.
- Singh SP. Integrated genetic improvement. In: Singh SP, editor. Common bean improvement in the twenty-first century. Dordrecht: Springer; 1999. p. 133–165.
- Singh SP. Production and utilization. In: Singh SP, editor. Common bean improvement in the twenty-first century. Dordrecht: Springer; 1999. p. 1–24.
- Terán H, Lema M, Webster D, et al. 75 years of breeding pinto bean for resistance to diseases in the United States. Euphytica. 2009;167(3):341–351.
- Velez JJ, Bassett MJ, Beaver JS, et al. Inheritance of resistance to bean golden mosaic virus in common bean. J Am Soc Hortic Sci. 1998;123(4):628–631.
- Blair MW, Giraldo MC, Buendia HF, et al. Microsatellite marker diversity in common bean (Phaseolus vulgaris L.). Theor Appl Genet. 2006;113(1):100–109.
- Román-Avilés B, Kelly JD. Identification of quantitative trait loci conditioning resistance to Fusarium root rot in common bean. Crop Sci. 2005;45(5):1881–1890.
- Haghighi KR, Ascher PD. Fertile, intermediate hybrids between Phaseolus vulgaris and P. acutifolius from congruity backcrossing. Sex Plant Reprod. 1988;1(1):51–58.
- Mejía-Jiménez A, Muñoz C, Jacobsen HJ, et al. Interspecific hybridization between common and tepary beans: increased hybrid embryo growth, fertility, and efficiency of hybridization through recurrent and congruity backcrossing. Theor Appl Genet. 1994;88(3–4):324–331.
- Singh SP, Urrea CA. Inter-and intraracial hybridization and selection for seed yield in early generations of common bean, Phaseolus vulgaris L. Euphytica. 1995;81(2):131–137.
- Nadeem MA, Nawaz MA, Shahid MQ, et al. DNA molecular markers in plant breeding: current status and recent advancements in genomic selection and genome editing. Biotechnol Biotechnol Equip. 2018;32(2):261–285.
- Nodari RO, Tsai SM, Guzman P, et al. Toward an integrated linkage map of common bean. III. Mapping genetic factors controlling host-bacteria interactions. Genetics. 1993;134(1):341–350.
- Adam-Blondon AF, Sévignac M, Bannerot H, et al. SCAR, RAPD and RFLP markers linked to a dominant gene (Are) conferring resistance to anthracnose in common bean. Theor Appl Genet. 1994;88(6–7):865–870.
- Vlasova A, Capella-Gutiérrez S, Rendón-Anaya M, et al. Genome and transcriptome analysis of the Mesoamerican common bean and the role of gene duplications in establishing tissue and temporal specialization of genes. Genome Biol. 2016;17(1):1–18.
- Vallejos CE, Sakiyama NS, Chase CD. A molecular marker-based linkage map of Phaseolus vulgaris L. Genetics. 1992;131(3):733–740.
- Yu K, Park SJ, Poysa V. Abundance and variation of microsatellite DNA sequences in beans (Phaseolus and Vigna). Genome. 1999;42(1):27–34.
- Yu K, Park SJ, Poysa V, et al. Integration of simple sequence repeat (SSR) markers into a molecular linkage map of common bean (Phaseolus vulgaris L.). J Hered. 2000;91(6):429–434.
- Guerra-Sanz JM. New SSR markers of Phaseolus vulgaris from sequence databases. Plant Breed. 2004;123(1):87–89.
- Garcia RA, Rangel PN, Brondani C, et al. The characterization of a new set of EST-derived simple sequence repeat (SSR) markers as a resource for the genetic analysis of Phaseolus vulgaris. BMC Genet. 2011;12(1):41–14.
- Hanai LR, De Campos T, Camargo LEA, et al. Development, characterization, and comparative analysis of polymorphism at common bean SSR loci isolated from genic and genomic sources. Genome. 2007;50(3):266–277.
- Caixeta ET, Borém A, Kelly JD. Development of microsatellite markers based on BAC common bean clones. Crop Breed Appl Biotechnol. 2005;5(2):125–133.
- Ragupathy R, Rathinavelu R, Cloutier S. Physical mapping and BAC-end sequence analysis provide initial insights into the flax (Linum usitatissimum L.) genome. BMC Genom. 2011;12(1):1–17.
- Schlueter JA, Goicoechea JL, Collura K, et al. BAC-end sequence analysis and a draft physical map of the common bean (Phaseolus vulgaris L.) genome. Trop Plant Biol. 2008;1(1):40–48.
- Yu K. Bacterial artificial chromosome libraries of pulse crops: characteristics and applications. J Biomed Biotechnol. 2012;2012:1–8.
- Phan NT, Sim SC. Genomic tools and their implications for vegetable breeding. Hortic Sci Technol. 2017;35(2):149–164.
- Chung YS, Choi SC, Jun TH, et al. Genotyping-by-sequencing: a promising tool for plant genetics research and breeding. Hortic Environ Biotechnol. 2017;58(5):425–431.
- Blair MW, Cortés AJ, Penmetsa RV, et al. A high-throughput SNP marker system for parental polymorphism screening, and diversity analysis in common bean (Phaseolus vulgaris L.). Theor Appl Genet. 2013;126(2):535–548.
- Song Q, Jia G, Hyten DL, et al. SNP assay development for linkage map construction, anchoring whole-genome sequence, and other genetic and genomic applications in common bean. G3 Genes Genomes Genet. 2015;5(11):2285–2290.
- Cichy KA, Wiesinger JA, Mendoza FA. Genetic diversity and genome-wide association analysis of cooking time in dry bean (Phaseolus vulgaris L.). Theor Appl Genet. 2015;128(8):1555–1567.
- Cichy KA, Porch TG, Beaver JS, et al. A Phaseolus vulgaris diversity panel for Andean bean improvement. Crop Sci. 2015;55(5):2149–2160.
- Kamfwa K, Cichy KA, Kelly JD. Genome-wide association study of agronomic traits in common bean. Plant Genome, 2015;8(2), 1–12.
- Hart JP, Griffiths PD. Genotyping-by-sequencing enabled mapping and marker development for the By-2 potyvirus resistance allele in common bean. Plant Genome. 2015;8(1):plantgenome2014-09.
- Diaz LM, Ricaurte J, Tovar E, et al. QTL analyses for tolerance to abiotic stresses in a common bean (Phaseolus vulgaris L.) population. PLoS ONE. 2018;13(8):e0202342.
- Miklas PN, Delorme R, Stone V, et al. Bacterial, fungal, and viral disease resistance loci mapped in a recombinant inbred common bean population (Dorado’/XAN 176). J Am Soc Hortic Sci. 2000;125(4):476–481.
- Miklas PN, Johnson WC, Delorme R, et al. QTL conditioning physiological resistance and avoidance to white mold in dry bean. Crop Sci. 2001;41(2):309–315.
- Miklas PN, Coyne DP, Grafton KF, et al. A major QTL for common bacterial blight resistance derives from the common bean great northern landrace cultivar Montana. Euphytica. 2003;131(1):137–146.
- Park SO, Coyne DP, Steadman JR, et al. Mapping of QTL for resistance to white mold disease in common bean. Crop Sci. 2001;41(4):1253–1262.
- McClean PE, Lee RK, Otto C, et al. Molecular and phenotypic mapping of genes controlling seed coat pattern and color in common bean (Phaseolus vulgaris L.). J Hered. 2002;93(2):148–152.
- Kelly JD, Gepts P, Miklas PN, et al. Tagging and mapping of genes and QTL and molecular marker-assisted selection for traits of economic importance in bean and cowpea. Field Crops Res. 2003;82(2–3):135–154.
- Kolkman JM, Kelly JD. QTL conferring resistance and avoidance to white mold in common bean. Crop Sci. 2003;43(2):539–548.
- López CE, Acosta IF, Jara C, et al. Identifying resistance gene analogs associated with resistances to different pathogens in common bean. Phytopathology. 2003;93(1):88–95.
- Papa R, Acosta J, Delgado-Salinas A, et al. A genome-wide analysis of differentiation between wild and domesticated Phaseolus vulgaris from Mesoamerica. Theor Appl Genet. 2005;111(6):1147–1158.
- Papa R, Bellucci E, Rossi M, et al. Tagging the signatures of domestication in common bean (Phaseolus vulgaris) by means of pooled DNA samples. Ann Bot. 2007;100(5):1039–1051.
- Felicetti E, Song Q, Jia G, et al. Simple sequence repeats linked with slow darkening trait in pinto bean discovered by single nucleotide polymorphism assay and whole genome sequencing. Crop Sci. 2012;52(4):1600–1608.
- Goretti D, Bitocchi E, Bellucci E, et al. Development of single nucleotide polymorphisms in Phaseolus vulgaris and related Phaseolus spp. Mol Breed. 2014;33(3):531–544.
- Hyten DL, Song Q, Fickus EW, et al. High-throughput SNP discovery and assay development in common bean. BMC Genom. 2010;11(1):475–478.
- Zou X, Shi C, Austin RS, et al. Genome-wide single nucleotide polymorphism and insertion-deletion discovery through next-generation sequencing of reduced representation libraries in common bean. Mol Breed. 2014;33(4):769–778.
- De Ron AM, Papa R, Bitocchi E, et al. Common bean. In De Ron AM, editor. Grain legumes. New York (NY): Springer; 2015. p. 1–36
- Assefa T, Mahama AA, Brown AV, et al. A review of breeding objectives, genomic resources, and marker-assisted methods in common bean. Mol Breed. 2019;39(2):20.
- Singh SP, Schwartz HF. Breeding common bean for resistance to diseases: a review. Crop Sci. 2010;50(6):2199–2223.
- Palacioglu G, Bayraktar H, Ozer G. Genetic variability of Colletotrichum lindemuthianum isolates from Turkey and resistance of Turkish bean cultivars. Span J Agric Res. 2020;18(3):e1005.
- Palacioglu G, Şanli İ, Bayraktar H, et al. Determination of Resistance Sources to BCMV and BCMNV in Some Common Bean (Phaseolus vulgaris L.) Cultivars Grown in Turkey. Int J Agric Wildl Sci. 2020;6(3):453–460.
- Yeken MZ, Özer G, ÇeliK A, et al. Identification of genes related to resistance for bean common mosaic virus and bean common mosaic necrosis virus in commercial common bean cultivars in Turkey. Turk J Agric Nat Sci. 2018;5(4):613–619.
- Ender M, Kelly JD. Identification of QTL associated with white mold resistance in common bean. Crop Sci. 2005;45(6):2482–2490.
- Mkwaila W, Terpstra KA, Ender M, et al. Identification of QTL for agronomic traits and resistance to white mold in wild and landrace germplasm of common bean. Plant Breed. 2011;130(6):665–672.
- Soule M, Porter L, Medina J, et al. Comparative QTL map for white mold resistance in common bean, and characterization of partial resistance in dry bean lines VA19 and I9365-3. Crop Sci. 2011;51(1):123–139.
- Oblessuc PR, Baroni RM, Garcia AAF, et al. Mapping of angular leaf spot resistance QTL in common bean (Phaseolus vulgaris L.) under different environments. BMC Genet. 2012;13(1):50–59.
- Keller B, Manzanares C, Jara C, et al. Fine-mapping of a major QTL controlling angular leaf spot resistance in common bean. Theor Appl Genet. 2015;128(5):813–826.
- Wang W, Jacobs JL, Chilvers MI, et al. QTL analysis of Fusarium root rot resistance in an Andean × middle American common bean RIL population. Crop Sci. 2018;58(3):1166–1180.
- Frei A, Blair MW, Cardona C, et al. QTL mapping of resistance to Thrips palmi Karny in common bean. Crop Sci. 2005;45(1):cropsci2005.0379-387.
- Kamfwa K, Beaver JS, Cichy KA, et al. QTL mapping of resistance to bean weevil in common bean. Crop Sci. 2018;58(6), 2370–2378.
- Dramadri IO, Nkalubo ST, Kelly JD. Identification of QTL associated with drought tolerance in Andean common bean. Crop Sci. 2019;59(3):1007–1020.
- Sedlar A, Zupin M, Maras M, et al. QTL mapping for drought-responsive agronomic traits associated with physiology, phenology, and yield in an andean intra-gene pool common bean population. Agronomy. 2020;10(2):225.
- Vallejo V, Kelly JD. 2002. The use of AFLP analysis to tag the Co-12 gene conditioning resistance to bean anthracnose In Proceedings of the X conference on plant and animal genome. http://www.intl-pag.org/pag/10/abstracts/PAGX_P233.html.
- Young RA, Melotto M, Nodari RO, et al. Marker-assisted dissection of the oligogenic anthracnose resistance in the common bean cultivar,’G2333. Theor Appl Genet. 1998;96(1):87–94.
- Trabanco N, Campa A, Ferreira JJ. Identification of a new chromosomal region involved in the genetic control of resistance to anthracnose in common bean. Plant Genome. 2015;8(2);plantgenome2014-10.
- Pérez-Vega E, Pañeda A, Rodríguez-Suárez C, et al. Mapping of QTLs for morpho-agronomic and seed quality traits in a RIL population of common bean. Theor Appl Genet. 2010;120(7):1367–1380.
- Méndez-Vigo B, Rodríguez-Suárez C, Paneda A, et al. Molecular markers and allelic relationships of anthracnose resistance gene cluster B4 in common bean. Euphytica. 2005;141(3):237–245.
- De Queiroz VT, De Sousa CS, Costa MR, et al. Development of SCAR markers linked to common bean angular leaf spot resistance genes. Annu Rep Bean Improv Coop. 2004;47:237–238.
- Awale HE, Kelly JD. Development of SCAR markers linked to Co-4^ 2 gene in common bean. Annual Report-Bean Improvement Cooperative, 44, 119–120. 2001.
- Campa A, Giraldez R, Ferreira JJ. Genetic dissection of the resistance to nine anthracnose races in the common bean differential cultivars MDRK and TU. Theor Appl Genet. 2009;119(1):1–11.
- Young RA, Kelly JD. RAPD markers linked to three major anthracnose resistance genes in common bean. Crop Sci. 1997;37(3):940–946.
- Campa A, Rodríguez-Suárez C, Pañeda A, et al. The bean anthracnose resistance gene Co-5, is located in linkage group B7. Annu Rep Bean Improv Coop. 2005;48:68.
- Geffroy V, Sicard D, de Oliveira JC, et al. Identification of an ancestral resistance gene cluster involved in the coevolution process between Phaseolus vulgaris and its fungal pathogen Colletotrichum lindemuthianum. Mol Plant Microbe Interact. 1999;12(9):774–784.
- Alzate-Marin AL, Costa MR, Arruda KM, et al. Characterization of the anthracnose resistance gene present in Ouro Negro (Honduras 35) common bean cultivar. Euphytica. 2003;133(2):165–169.
- Melotto M, Kelly J. SCAR markers linked to major disease resistance genes in common bean. Annu Rep Bean Improv Coop. 1998;41:64–65.
- Haley SD, Afanador LK, Miklas PN, et al. Heterogeneous inbred populations are useful as sources of near-isogenic lines for RAPD marker localization. Theor Appl Genet. 1994;88(3):337–342.
- Miklas PN. Marker-assisted selection for disease resistance in common bean. Annu Rep Bean Improv Coop. 2002;45:1–3.
- Park SO, Coyne DP, Steadman JR. Confirmation of the Ur-6 location in Phaseolus vulgaris L. Annu Rep Bean Improv Coop. 2004;47:265–266.
- Park SO, Coyne DP, Steadman JR, et al. Mapping of the Ur-7 gene for specific resistance to rust in common bean. Crop Sci. 2003;43(4):1470–1476.
- Jung G, Coyne DP, Bokosi J, et al. Mapping genes for specific and adult plant resistance to rust and abaxial leaf pubescence and their genetic relationships using randomly amplified polymorphic DNA (RAPD) markers in common bean. J Am Soc Hort Sci. 1998;123(5):859–863.
- Park SO, Coyne DP, Bokosi JM, et al. Molecular markers linked to genes for specific rust resistance and indeterminate growth habit in common bean. Euphytica. 1999;105(2):133–141.
- Awale HE, Safeena MI, Vallejo VA, et al. Sq4 Scar Marker Linked To The Co-2 Gene On Bll Appears To Be Linked To The Ur-11 Gene. 2008.
- Johnson E, Miklas PN, Stavely JR, et al. Coupling-and repulsion-phase RAPDs for marker-assisted selection of PI 181996 rust resistance in common bean. Theor Appl Genet. 1995;90(5):659–664.
- Mienie CMS, Liebenberg MM, Pretorius ZA, et al. SCAR markers linked to the common bean rust resistance gene Ur-13. Theor Appl Genet. 2005;111(5):972–979.
- Melotto M, Afanador L, Kelly JD. Development of a SCAR marker linked to the I gene in common bean. Genome. 1996;39(6):1216–1219.
- Strausbaugh CA, Myers JR, Forster RL, et al. bc-1 and bc-u—two loci controlling bean common mosaic virus resistance in common bean are linked. J Am Soc Hortic Sci. 1999;124(6):644–648.
- Johnson WC, Guzmán P, Mandala D, et al. Molecular tagging of the bc-3 gene for introgression into Andean common bean. Crop Sci. 1997;37(1):248–254.
- Mukeshimana G, Paneda A, Rodríguez-Suárez C, et al. Markers linked to the bc-3 gene conditioning resistance to bean common mosaic potyviruses in common bean. Euphytica. 2005;144(3):291–299.
- Urrea CA, Miklas PN, Beaver JS, et al. A codominant randomly amplified polymorphic DNA (RAPD) marker useful for indirect selection of bean golden mosaic virus resistance in common bean. J Am Soc Hortic Sci. 1996;121(6):1035–1039.
- Blair MW, Beaver JS, Nin JC, et al. Registration of PR9745-232 and RMC-3 red-mottled dry bean germplasm lines with resistance to Bean golden yellow mosaic virus. Crop Sci. 2006;46(2):1000–1002.
- Chen J, Zhang X, Jing R, et al. Cloning and genetic diversity analysis of a new P5CS gene from common bean (Phaseolus vulgaris L.). Theor Appl Genet. 2010;120(7):1393–1404.
- Blair MW, Galeano CH, Tovar E, et al. Development of a Mesoamerican intra-genepool genetic map for quantitative trait loci detection in a drought tolerant × susceptible common bean (Phaseolus vulgaris L.) cross. Mol Breed. 2012;29(1):71–88.
- Johannsen W. The genotype conception of heredity. Am Nat. 1911;45(531):129–159.
- Motto M, Soressi GP, Salamini F. Seed size inheritance in a cross between wild and cultivated common beans (Phaseolus vulgaris L.). Genetica. 1978;49(1):31–36.
- Vallejos CE, Chase CD. Linkage between isozyme markers and a locus affecting seed size in Phaseolus vulgaris L. Theor Appl Genet. 1991;81(3):413–419.
- Blair MW, Izquierdo P. Use of the advanced backcross-QTL method to transfer seed mineral accumulation nutrition traits from wild to Andean cultivated common beans. Theor Appl Genet. 2012;125(5):1015–1031.
- Koinange EM, Singh SP, Gepts P. Genetic control of the domestication syndrome in common bean. Crop Sci. 1996;36(4):1037–1045.
- Tar’an B, Michaels TE, Pauls KP. Genetic mapping of agronomic traits in common bean. Crop Sci. 2002;42(2):544–556.
- Checa OE, Blair MW. Mapping QTL for climbing ability and component traits in common bean (Phaseolus vulgaris L.). Mol Breed. 2008;22(2):201–215.
- Garcia RAV, Rangel PN, Bassinello PZ, et al. QTL mapping for the cooking time of common beans. Euphytica. 2012;186(3):779–792.
- Park SO, Coyne DP, Jung G, et al. Mapping of QTL for seed size and shape traits in common bean. J Am Soc Horti Sci. 2000;125(4):466–475.
- Beebe SE, Rojas-Pierce M, Yan X, et al. Quantitative trait loci for root architecture traits correlated with phosphorus acquisition in common bean. Crop Sci. 2006;46(1):413–423.
- Hoyos-Villegas V, Song Q, Wright EM, et al. Joint linkage QTL mapping for yield and agronomic traits in a composite map of three common bean RIL populations. Crop Sci. 2016;56(5):2546–2563.
- Bhakta MS, Gezan SA, Clavijo Michelangeli JA, et al. A predictive model for time-to-flowering in the common bean based on QTL and environmental variables. G3 (Bethesda). 2017;7(12):3901–3912.
- Sandhu KS, You FM, Conner RL, et al. Genetic analysis and QTL mapping of the seed hardness trait in a black common bean (Phaseolus vulgaris) recombinant inbred line (RIL) population. Mol Breed. 2018;38(3):1–13.
- Lei L, Wang L, Wang S, et al. Marker-trait association analysis of seed traits in accessions of common bean (Phaseolus vulgaris L.) in China. Front Genet. 2020;11:698.
- Geravandi M, Cheghamirza K, Farshadfar E, et al. QTL analysis of seed size and yield-related traits in an inter-genepool population of common bean (Phaseolus vulgaris L.). Sci Hortic. 2020;274:109678.
- Blair MW, Knewtson SJ, Astudillo C, et al. Variation and inheritance of iron reductase activity in the roots of common bean (Phaseolus vulgaris L.) and association with seed iron accumulation QTL. BMC Plant Biol. 2010;10(1):215–212.
- Nemli S, Asciogul TK, Ates D, Esiyok, et al. SNP identification through genotyping by sequencing and genome-wide association study (GWAS) of pod traits in common bean. In International Plant and Animal Genome Conference, Paper. 2016; Vol. 19798).
- Moghaddam SM, Mamidi S, Osorno JM, et al. Genome-wide association study identifies candidate loci underlying agronomic traits in a Middle American diversity panel of common bean. Plant Genome. 2016;9(3):1–21.
- Perseguini JMKC, Oblessuc PR, Rosa JRBF, et al. Genome-wide association studies of anthracnose and angular leaf spot resistance in common bean (Phaseolus vulgaris L.). PLoS ONE. 2016;11(3):e0150506.
- Ferreira JJ, Murube E, Campa A. Introgressed genomic regions in a set of near-isogenic lines of common bean revealed by genotyping-by-sequencing. Plant Genome. 2016;10(1):plantgenome2016-08.
- Berny Mier Y, Teran JC, Konzen ER, et al. Root and shoot variation in relation to potential intermittent drought adaptation of Mesoamerican wild common bean (Phaseolus vulgaris L. Ann Bot. 2019;124(6):917–932.
- Emiliano VP, González-Chavira Mario M, Patricia GC, et al. Identification of novel drought-tolerant-associated SNPs in common bean (Phaseolus vulgaris). Front Plant Sci. 2015;6:546.
- Hoyos-Villegas V, Song Q, Kelly JD. Genome-wide association analysis for drought tolerance and associated traits in common bean. Plant Genome. 2017;10(1):1–17.
- Wu L, Chang Y, Wang L, et al. Genetic dissection of drought resistance based on root traits at the bud stage in common bean. Theor Appl Genet. 2021;134:1–15.
- Leitão ST, Bicho MC, Pereira P, et al. Common bean SNP alleles and candidate genes affecting photosynthesis under contrasting water regimes. Hortic Res. 2021;8(1):1–14.
- Nemli S, Asciogul TK, Kaya HB, et al. Association mapping for five agronomic traits in the common bean (Phaseolus vulgaris L.). J Sci Food Agric. 2014;94(15):3141–3151.
- Zuiderveen GH, Padder BA, Kamfwa K, et al. Genome-wide association study of anthracnose resistance in Andean beans (Phaseolus vulgaris). PLoS ONE. 2016;11(6):e0156391.]
- Tock AJ, Fourie D, Walley PG, et al. Genome-wide linkage and association mapping of halo blight resistance in common bean to race 6 of the globally important bacterial pathogen. Front Plant Sci. 2017;8:1170.
- Wu C, Chen Z, Dardalhon V, et al. The transcription factor musculin promotes the unidirectional development of peripheral Treg cells by suppressing the TH2 transcriptional program. Nat Immunol. 2017;18(3):344–353.
- Wu J, Chen J, Wang L, et al. Genome-wide investigation of WRKY transcription factors involved in terminal drought stress response in common bean. Front Plant Sci. 2017;8:380.
- Ates D, Asciogul TK, Nemli S, et al. Association mapping of days to flowering in common bean (Phaseolus vulgaris L.) revealed by DArT markers. Mol Breed. 2018;38(9):113.
- Katuuramu DN, Hart JP, Porch TG, et al. Genome-wide association analysis of nutritional composition-related traits and iron bioavailability in cooked dry beans (Phaseolus vulgaris L.). Mol Breed. 2018;38(4):44.
- Nascimento M, Nascimento ACC, Silva FFE, et al. Quantile regression for genome-wide association study of flowering time-related traits in common bean. PLoS ONE. 2018;13(1):e0190303.
- Oladzad A, Porch T, Rosas JC, et al. Single and multi-trait GWAS identify genetic factors associated with production traits in common bean under abiotic stress environments. G3 (Bethesda)). 2019;9(6):1881–1892.
- Tigist SG, Melis R, Sibiya J, et al. Population structure and genome-wide association analysis of bruchid resistance in Ethiopian common bean genotypes. Crop Sci. 2019;59(4):1504–1515.
- Raggi L, Caproni L, Carboni A, et al. Genome-wide association study reveals candidate genes for flowering time variation in common bean (Phaseolus vulgaris L.). Front Plant Sci. 2019;10:962.
- Jain S, Poromarto S, Osorno JM, et al. Genome wide association study discovers genomic regions involved in resistance to soybean cyst nematode (Heterodera glycines) in common bean. PLoS ONE. 2019;14(2):e0212140.
- Wu D, Hought J, Baseggio M, et al. Genomic characterization of the native seeds/search common bean (Phaseolus vulgaris L.) Collection and its seed coat patterns. Genet Resour Crop Evol. 2019;66(7):1469–1482.
- Wen L, Chang HX, Brown PJ, et al. Genome-wide association and genomic prediction identifies soybean cyst nematode resistance in common bean including a syntenic region to soybean Rhg1 locus. Hortic Res. 2019;6(1):9–12.
- Ojwang PPO, Eldridge T, Corredor-Moreno P, et al. Genome-wide association study of resistance to bean fly and population structure of market classes of common bean. BioRxiv, 633545. 2019.
- López-Hernández F, Cortés AJ. Last-generation genome-environment associations reveal the genetic basis of heat tolerance in common bean (Phaseolus vulgaris L.). Front Genet. 2019;10:954.
- Myers JR, Wallace LT, Mafi Moghaddam S, et al. Improving the health benefits of snap bean: genome-wide association studies of total phenolic content. Nutrients. 2019;11(10):2509.
- Caproni L, Raggi L, Talsma EF, et al. European landrace diversity for common bean biofortification: a genome-wide association study. Sci Rep. 2020;10(1):1–13.
- Erdogmus S, Ates D, Nemli S, et al. Genome-wide association studies of Ca and Mn in the seeds of the common bean (Phaseolus vulgaris L.). Genomics. 2020;112(6):4536–4546.
- Banoo A, Nabi A, Rasool RS, et al. North-Western Himalayan Common Beans: Population Structure and Mapping of Quantitative Anthracnose Resistance Through Genome Wide Association Study. Front Plant Sci. 2020;11:1459.
- Vidigal Filho PS, Gonçalves-Vidigal MC, Vaz Bisneta M, et al. Genome-wide association study of resistance to anthracnose and angular leaf spot in Brazilian Mesoamerican and Andean common bean cultivars. Crop Sci. 2020;60(6):2931–2950.
- Campa A, García-Fernández C, Ferreira JJ. Genome-wide association study (GWAS) for resistance to Sclerotinia sclerotiorum in common bean. Genes. 2020;11(12):1496.
- Dramadri IO, Amongi W, Kelly JD, et al. Genome-wide association analysis of resistance to Pythium ultimum in common bean (Phaseolus vulgaris. Plant Breed. 2020;139(6):1168–1180.
- Maldonado-Mota CR, Moghaddam SM, Schröder S, et al. Genomic regions associated with resistance to anthracnose in the Guatemalan climbing bean (Phaseolus vulgaris L.) germplasm collection. Genet Resour Crop Evol. 2020;68:1–11.
- Costa LC, Nalin RS, Dias MA, et al. Different loci control resistance to different isolates of the same race of Colletotrichum lindemuthianum in common bean. Theor Appl Genet. 2021;1–14.
- Semagn K, Babu R, Hearne S, et al. Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): overview of the technology and its application in crop improvement. Mol Breed. 2014;33(1):1–14.
- Steele KA, Quinton-Tulloch MJ, Amgai RB, et al. Accelerating public sector rice breeding with high-density KASP markers derived from whole genome sequencing of indica rice. Mol Breed. 2018;38(4):38–13.
- Hurtado-Gonzales OP, Valentini G, Gilio TAS, et al. Fine mapping of genes conferring resistance to rust and anthracnose of common bean. Annu Rep Bean Improv Coop. 2017;61:27–28.
- Gilio TAS, Hurtado-Gonzales OP, Gonçalves-Vidigal MC, et al. Fine mapping of an anthracnose-resistance locus in Andean common bean cultivar Amendoim Cavalo. PLoS ONE. 2020;15(10):e0239763.
- Libault M, Dickstein R. Advances in functional genomics in legumes. In: Gupta S, Nadarajan N, Gupta DS, editors. Legumes in the Omic Era. New York (NY): Springer; 2014. p. 15–39.
- Wang X, Elling AA, Li X, et al. Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell. 2009;21(4):1053–1069.
- Hatey F, Tosser-Klopp G, Clouscard-Martinato C, et al. Expressed sequence tags for genes: a review. Genet Sel Evol. 1998;30(6):521–541.
- Asamizu E, Nakamura Y, Sato S, et al. Characteristics of the Lotus japonicus gene repertoire deduced from large-scale expressed sequence tag (EST) analysis. Plant Mol Biol. 2004;54(3):405–414.
- Blair MW, Fernandez AC, Ishitani M, et al. Construction and EST sequencing of full-length, drought stress cDNA libraries for common beans (Phaseolus vulgaris L.). BMC Plant Biol. 2011;11(1):171–115.
- Cheung F, Haas BJ, Goldberg SM, et al. Sequencing Medicago truncatula expressed sequenced tags using 454 Life Sciences technology. BMC Genom. 2006;7(1):1–10.
- Vodkin LO, Khanna A, Shealy R, et al. Microarrays for global expression constructed with a low redundancy set of 27,500 sequenced cDNAs representing an array of developmental stages and physiological conditions of the soybean plant. BMC Genom. 2004;5(1):73–18.
- Blair MW, Torres MM, Giraldo MC, et al. Development and diversity of Andean-derived, gene-based microsatellites for common bean (Phaseolus vulgaris L.). BMC Plant Biol. 2009;9(1):100–114.
- Melotto M, Monteiro-Vitorello CB, Bruschi AG, et al. Comparative bioinformatic analysis of genes expressed in common bean (Phaseolus vulgaris L.) seedlings. Genome. 2005;48(3):562–570.
- Ramírez M, Graham MA, Blanco-López L, et al. Sequencing and analysis of common bean ESTs. Building a foundation for functional genomics. Plant Physiol. 2005;137(4):1211–1227.
- Tian J, Venkatachalam P, Liao H, et al. Molecular cloning and characterization of phosphorus starvation responsive genes in common bean (Phaseolus vulgaris L.). Planta. 2007;227(1):151–165.
- McClean PE, Mamidi S, McConnell M, et al. Synteny mapping between common bean and soybean reveals extensive blocks of shared loci. BMC Genom. 2010;11(1):184–110.
- Blair MW, Hurtado N, Chavarro CM, et al. Gene-based SSR markers for common bean (Phaseolus vulgaris L.) derived from root and leaf tissue ESTs: an integration of the BMc series. BMC Plant Biol. 2011;11(1):50–10.
- Thibivilliers S, Joshi T, Campbell KB, et al. Generation of Phaseolus vulgaris ESTs and investigation of their regulation upon Uromyces appendiculatus infection. BMC Plant Biol. 2009;9(1):46–13.
- Kalavacharla V, Liu Z, Meyers BC, et al. Identification and analysis of common bean (Phaseolus vulgaris L.) transcriptomes by massively parallel pyrosequencing. BMC Plant Biol. 2011;11(1):135–118.
- Kavas M, Kizildogan A, Gökdemir G, et al. Genome-wide investigation and expression analysis of AP2-ERF gene family in salt tolerant common bean. EXCLI J. 2015;14:1187–1206.
- Kavas M, Baloğlu MC, Atabay ES, et al. Genome-wide characterization and expression analysis of common bean bHLH transcription factors in response to excess salt concentration. Mol Genet Genom. 2016;291(1):129–143.
- Büyük İ . Inal B, Ilhan E, Tanriseven M, Aras S, Erayman M. Genome-wide identification of salinity responsive HSP70s in common bean. Mol Biol Rep. 2016;43(11):1251–1266.
- Wu J, Wang L, Wang S. Comprehensive analysis and discovery of drought-related NAC transcription factors in common bean. BMC Plant Biol. 2016;16(1):1–13.
- İnal B, Büyük İ, İlhan E, et al. Genome-wide analysis of Phaseolus vulgaris C2C2-YABBY transcription factors under salt stress conditions. 3 Biotech. 2017;7(5):302.
- Ilhan E. Genome-Wide Characterization and Analysis of Sbp Transcription Factor Family in Common Bean (Phaseolus vulgaris L. Appl Ecol Env Res. 2018;16(5):5467–5480.
- Buyuk I, Inal B, Ilhan E, et al. Common Bean (Phaseolus vulgaris L.) Dof Transcription Factors Differentially Expressed Under Salt Stress. Commun Fac Sci Univ Ankara Ser C Biol. 2019;28(1):43–66.
- Gökdemir FS. Genome-Wide Identification and Characterization of Whirly and Arr-B Genes in Common Bean (Phaseolus vulgaris L.) and Assessment of Their Possible Role in Biotic Stress Response at Gene Expression Level, Ph.D. Thesis, Ankara University, Graduate School of Natural and Applied Science, Department of Biology. 2019.
- Zhang Q, Li WJ, Zhang WJ, et al. PvB3s: genome-wide and transcriptome-wide identification and analysis reveals the B3 family regulate auxin to resist salt stress at the sprout stage in common bean (P. vulgaris L.). 2020. https://doi.org/10.21203/rs.3.rs-36056/v1.
- Silva DAD, Tsai SM, Chiorato AF, et al. Analysis of the common bean (Phaseolus vulgaris L.) transcriptome regarding efficiency of phosphorus use. PLoS ONE. 2019;14(1):e0210428.
- Hernández G, Ramírez M, Valdés-López O, et al. Phosphorus stress in common bean: root transcript and metabolic responses. Plant Physiol. 2007;144(2):752–767.
- Bliss FA, Brown JW. Breeding common bean for improved quantity and quality of seed protein. In: Janick J, editor. Plant breeding reviews. Boston (MA): Springer; 1983. p. 59–102.
- FAO. Food and Agriculture Organization of the United Nations. 2013. Available from http://www.fao.org/faostat/en/#data/TP.
- Vitale A, Bollini R. Legume storage proteins. In: Kigel J, Gad G, editors. Seed Development and Germination. Boca Raton: Routledge; 1995. p. 73–102.
- Mühling M, Gilroy J, Croy RR. Legumin proteins from seeds of Phaseolus vulgaris L. J Plant Physiol. 1997;150(4):489–492.
- Gepts P, Bliss FA. Enhanced available methionine concentration associated with higher phaseolin levels in common bean seeds. Theor Appl Genet. 1984;69(1):47–53.
- Osborn TC, Bliss FA. Effects of genetically removing lectin seed protein on horticultural and seed characteristics of common bean. J Am Soc Hortic Sci. 1985;110:484–488.
- Voelker TA, Staswick P, Chrispeels MJ. Molecular analysis of two phytohemagglutinin genes and their expression in Phaseolus vulgaris cv. Pinto, a lectin-deficient cultivar of the bean. Embo J. 1986;5(12):3075–3082.
- Bollini R, Ceriotti A, Daminati MG, et al. Glycosylation is not needed for the intracellular transport of phytohemagglutinin in developing Phaseolus vulgaris cotyledons and for the maintenance of its biological activities. Physiol Plant. 1985;65(1):15–22.
- De La Fuente M, Borrajo A, Bermúdez J, et al. 2-DE-based proteomic analysis of common bean (Phaseolus vulgaris L.) seeds. J Proteom. 2011;74(2):262–267.
- Robison FM, Heuberger AL, Brick MA, et al. Proteome characterization of leaves in common bean. Proteomes. 2015;3(3):236–248.
- Torres NL, Cho K, Shibato J, et al. Gel-based proteomics reveals potential novel protein markers of ozone stress in leaves of cultivated bean and maize species of Panama. Electrophoresis. 2007;28(23):4369–4381.
- Zadražnik T, Hollung K, Egge-Jacobsen W, et al. Differential proteomic analysis of drought stress response in leaves of common bean (Phaseolus vulgaris L.). J Proteomics. 2013;78:254–272.
- Parreira JR, Bouraada J, Fitzpatrick MA, et al. Differential proteomics reveals the hallmarks of seed development in common bean (Phaseolus vulgaris L.). J Proteomics. 2016;143:188–198.
- Badowiec A, Weidner S. Proteomic changes in the roots of germinating Phaseolus vulgaris seeds in response to chilling stress and post-stress recovery. J Plant Physiol. 2014;171(6):389–398.
- Salavati A, Taleei A, Akbar Shahnejat Bushehri A, et al. Analysis of the proteome of common bean (Phaseolus vulgaris L.) roots after inoculation with Rhizobium etli. Protein Pept Lett. 2012;19(8):880–889.
- Lee J, Feng J, Campbell KB, et al. Quantitative proteomic analysis of bean plants infected by a virulent and avirulent obligate rust fungus. Mol Cell Proteom. 2009;8(1):19–31.
- Natarajan SS, Pastor-Corrales MA, Khan FH, et al. Proteomic analysis of common bean (Phaseolus vulgaris L.) by two-dimensional gel electrophoresis and mass spectrometry. J Basic Appl Sci. 2013;9:424–437.
- López-Pedrouso M, Alonso J, Zapata C. Evidence for phosphorylation of the major seed storage protein of the common bean and its phosphorylation-dependent degradation during germination. Plant Mol Biol. 2014;84(4–5):415–428.
- Mensack MM, Fitzgerald VK, Ryan EP, et al. Evaluation of diversity among common beans (Phaseolus vulgaris L.) from two centers of domestication using’omics’ technologies. BMC Genom. 2010;11(1):1–11.
- Lei Z, Dai X, Watson BS, et al. A legume specific protein database (LegProt) improves the number of identified peptides, confidence scores and overall protein identification success rates for legume proteomics. Phytochemistry. 2011;72(10):1020–1027.
- El-Gebali S, Mistry J, Bateman A, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47(D1):D427–D432.
- Guo AY, Zhu QH, Chen X, et al. GSDS: a gene structure display server. Yi Chuan. 2007;29(8):1023–1026.
- Bailey TL, Williams N, Misleh C, et al. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34(Web Server issue):W369–W373.
- Horton P, Park KJ, Obayashi T, et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35(Web Server issue):W585–W587.
- Emanuelsson O, Brunak S, Von Heijne G, et al. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2(4):953–971.
- Zhang Y. miRU: an automated plant miRNA target prediction server. Nucleic Acids Res. 2005;33(Web Server issue):W701–W704.
- Conesa A, Götz S, García-Gómez JM, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676.
- Lee TH, Tang H, Wang X, et al. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 2013;41(Database issue):D1152–D1158.
- Zheng Y, Jiao C, Sun H, et al. iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol Plant. 2016;9(12):1667–1670.
- Kelley LA, Mezulis S, Yates CM, et al. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10(6):845–858.
- Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34(Web Server issue):W609–W612.
- Gautam B, & Bonthala VS, Gajula MP. PvTFDB: a Phaseolus vulgaris transcription factors database for expediting functional genomics in legumes. DATABASE-OXFORD. 2016;2016:baw114.
- Caldas DGG, Konzen ER, Recchia GH, et al. Functional genomics of biotic and abiotic stresses in Phaseolus vulgaris. In: Shanker AK, Shanker C, editor. Abiotic and biotic stress in plants-recent advances and future perspectives. London: IntechOpen; 2016. p. 121–150.
- Amugune NO, Anyango B, Mukiama TK. Arobacterium-mediated transformation of common bean. Afr Crop Sci J. 2011;19(3):137–147.
- Aragão FJ, Vianna GR, Albino MM, et al. Transgenic dry bean tolerant to the herbicide glufosinate ammonium. Crop Sci. 2002;42(4):1298–1302.
- Dillen W, Engler G, Van Montagu M, et al. Electroporation-mediated DNA delivery to seedling tissues of Phaseolus vulgaris L.(common bean). Plant Cell Rep. 1995;15(1–2):119–124.
- Kim JW, Minamikawa T. Transformation and regeneration of French bean plants by the particle bombardment process. Plant Sci. 1996;117(1–2):131–138.
- Rech EL, Vianna GR, Aragao FJ. High-efficiency transformation by biolistics of soybean, common bean and cotton transgenic plants. Nat Protoc. 2008;3(3):410–418.
- Aragão FJ, de Sa MFG, Davey MR, et al. Factors influencing transient gene expression in bean (Phaseolus vulgaris L.) using an electrical particle acceleration device. Plant Cell Rep. 1993;12(9):483–490.
- Russell DR, Wallace KM, Bathe JH, et al. Stable transformation of Phaseolus vulgaris via electric-discharge mediated particle acceleration. Plant Cell Rep. 1993;12(3):165–169.
- Aragão FJL, Barros LMG, Brasileiro ACM, et al. Inheritance of foreign genes in transgenic bean (Phaseolus vulgaris L.) co-transformed via particle bombardment. Theor Appl Genet. 1996;93(1–2):142–150.
- Aragão FJ, Rech EL. Morphological factors influencing recovery of transgenic bean plants (Phaseolus vulgaris L.) of a carioca cultivar. Int J Plant Sci. 1997;158(2):157–163.
- Aragão FJL, Barros LMG, De Sousa MV, et al. Expression of a methionine-rich storage albumin from the Brazil nut (Bertholletia excelsa HBK, Lecythidaceae) in transgenic bean plants (Phaseolus vulgaris L., Fabaceae). Genet Mol Biol. 1999;22(3):445–449.
- Faria JC, Albino MM, Dias BB, et al. Partial resistance to Bean golden mosaic virus in a transgenic common bean (Phaseolus vulgaris L.) line expressing a mutated rep gene. Plant Sci. 2006;171(5):565–571.
- Bonfim K, Faria JC, Nogueira EO, et al. RNAi-mediated resistance to Bean golden mosaic virus in genetically engineered common bean (Phaseolus vulgaris). Mol Plant Microbe Interact. 2007;20(6):717–726.
- Aragão FJ, Nogueira EO, Tinoco MLP, et al. Molecular characterization of the first commercial transgenic common bean immune to the Bean golden mosaic virus. J Biotechnol. 2013;166(1–2):42–50.
- Aragão FJL, Ribeiro SG, Barros LMG, et al. Transgenic beans (Phaseolus vulgaris L.) engineered to express viral antisense RNAs show delayed and attenuated symptoms to bean golden mosaic geminivirus. Mol Breed. 1998;4(6):491–499.
- Franklin CI, Trieu TN, Cassidy BG, et al. Genetic transformation of green bean callus via Agrobacterium mediated DNA transfer. Plant Cell Rep. 1993;12(2):74–79.
- Zhang Z, Coyne DP, Mitra A. Factors affecting Agrobacterium-mediated transformation of common bean. J Am Soc Hortic Sci. 1997;122(3):300–305.
- Arellano J, Fuentes SI, Castillo-Espana P, et al. Regeneration of different cultivars of common bean (Phaseolus vulgaris L.) via indirect organogenesis. Plant Cell Tiss Organ Cult. 2009;96(1):11–18.
- Colpaert N, Tilleman S, Van Montagu M, et al. Composite Phaseolus vulgaris plants with transgenic roots as research tool. Afr J Biotechnol. 2008;7(4).
- Genga AM, Allavena A, Ceriotti A, et al. Genetic transformation in Phaseolus species by high-velocity microprojectiles. In Vitro Culture, XXIII IHC. 1990;300:309–314.
- McClean P, Chee P, Held B, et al. Susceptibility of dry bean (Phaseolus vulgaris L.) to Agrobacterium infection: transformation of cotyledonary and hypocotyl tissues. Plant Cell Tiss Organ Cult. 1991;24(2):131–138.
- Lewis ME, Bliss FA. Tumor formation and β-glucuronidase expression in Phaseolus vulgaris inoculated with Agrobacterium tumefaciens. J Am Soc Hortic Sci. 1994;119(2):361–366.
- Kapila J, De Rycke R, Van Montagu M, et al. An Agrobacterium-mediated transient gene expression system for intact leaves. Plant Sci. 1997;122(1):101–108.
- Mukeshimana G, Ma Y, Walworth AE, et al. Factors influencing regeneration and Agrobacterium tumefaciens-mediated transformation of common bean (Phaseolus vulgaris L.). Plant Biotechnol Rep. 2013;7(1):59–70.
- Estrada-Navarrete G, Alvarado-Affantranger X, Olivares JE, et al. Agrobacterium rhizogenes transformation of the Phaseolus spp.: a tool for functional genomics. Mol Plant Microbe Interact. 2006;19(12):1385–1393.
- Estrada-Navarrete G, Alvarado-Affantranger X, Olivares JE, et al. Fast, efficient and reproducible genetic transformation of Phaseolus spp. by Agrobacterium rhizogenes. Nat Protoc. 2007;2(7):1819–1824.
- Xue R, Wu X, Wang Y, et al. Hairy root transgene expression analysis of a secretory peroxidase (PvPOX1) from common bean infected by Fusarium wilt. Plant Sci. 2017;260:1–7.
- Liu Z, Park BJ, Kanno A, et al. The novel use of a combination of sonication and vacuum infiltration in Agrobacterium-mediated transformation of kidney bean (Phaseolus vulgaris L.) with lea gene. Mol Breed. 2005;16(3):189–197.
- Espinosa-Huerta E, Quintero-Jiménez A, Cabrera-Becerra K, et al. Stable and efficient Agrobacterium tumefaciens mediated transformation of Phaseolus vulgaris. Agrociencia. 2013;47(4):319–333.
- Dang W, Wei ZM. High frequency plant regeneration from the cotyledonary node of common bean. Biol Plant. 2009;53(2):312–316.
- Thảo NT, Thảo NT, Hassan F, Jacobsen HJ, et al. In vitro propagation of common bean (Phaseolus vulgaris L.). J Sci Dev. 2013;11:868–876.
- Castillo BM, Rodríguez de la O JL, Gallardo JOM, et al. In vitro plants of common bean (Phaseolus vulgaris L.) obtained by direct organogenesis. JAS. 2015;7(11):169.
- Quintero-Jiménez A, Espinosa-Huerta E, Acosta-Gallegos JA, et al. Enhanced shoot organogenesis and regeneration in the common bean (Phaseolus vulgaris L.). Plant Cell Tiss Organ Cult. 2010;102(3):381–386.
- Veltcheva M, Svetleva D. In vitro regeneration of Phaseolus vulgaris L. via organogenesis from petiole explants. J Cent Eur Agric. 2005;6(1):53–58.
- Sabzikar R, Sticklen MB, Kelly JD. In vitro regeneration and morphogenesis studies in common bean. Plant Cell, Tissue Organ Cult. 2010;100(1):97–105.
- Souter JR, Gurusamy V, Porch TG, et al. Successful introgression of abiotic stress tolerance from wild tepary bean to common bean. Crop Sci. 2017;57(3):1160–1171.
- Melotto M, Kelly JD. An allelic series at the Co-1 locus conditioning resistance to anthracnose in common bean of Andean origin. Euphytica. 2000;116(2):143–149.
- You MP, Colmer TD, Barbetti MJ. Salinity drives host reaction in Phaseolus vulgaris (common bean) to Macrophomina phaseolina. Funct Plant Biol. 2011;38(12):984–992.