Abstract
Alkaline stress has been considered a major environmental threat to plant growth and agricultural production. Even though the mechanisms have been characterized in some plants, the information about the alkaline stress-induced defense responses in soybean is insufficient. To identify defense related genes, the transcriptional profiles of soybean seedlings were examined using RNA sequencing under alkaline stress. The results showed that a total of 4603 genes were up-regulated and 1659 were down-regulated compared to expression in control plants. Further, the functional enrichment analysis showed that these differentially expressed genes were significantly enriched in 43 pathways and mainly associated with catalytic activity and binding, metabolic process, cellular process and cell progress. Then, 19 transcription factors from AP2-EREBF, MYB families and isoflavone related genes were chosen and identified to be induced under alkaline stress by quantitative real-time polymerase chain reaction (qRT-PCR). Taken together, our results highlight the differentially expressed genes and identify a set of genes associated with the response to alkaline stress, which findings establish a foundation for further analyses of soybean alkaline stress tolerance mechanisms.
Supplemental data for this article is available online at https://doi.org/10.1080/13102818.2021.1976078 .
Introduction
Alkaline stress has been considered as a major detrimental factor for crop productivity. A total of 3,105,000 km2 of global land is affected by alkaline soil [Citation1]. Compared with neutral salts stress such as NaCl and Na2SO4, alkaline stress due to NaHCO3 and Na2CO3 stress may cause more harmful effects to plant growth [Citation2]. Soil alkalinity inhibits plant growth and development by means of high pH, HCO3- and CO32-. Severe alkaline stress can inhibit sugar production, N metabolism and photosynthesis, as well as disrupt the ionic homeostasis and limits the absorption of ions, such as H2PO4-, Cl-, Al3+ and Fe2+ [Citation3, Citation4]. Previous studies have showed that high soil pH affects nutrient uptake, organic acids balance and ions balance [Citation5]. Also, the high pH could initiate the accumulation of malonic dialdehyde and reactive oxygen species [Citation6]. Therefore, alkaline stress is different from other types of abiotic stress, so plant cells may respond to it with a more complex tolerance mechanism [Citation7]. Thus, an understanding regarding the basic mechanisms of plant responses towards alkaline stress is urgently needed as little is known about basic alkaline stress responses especially in soybean crop.
Soybean has been adopted globally as one of the most important crops. Unlike other cereal crops, where carbohydrates are the main constituent of the nutrient profile, soybean is comprised of protein, oil, micronutrients and isoflavones and thus represents greater nutritive value [Citation8, Citation9]. The productivity of soybean is significantly hampered by abiotic stresses, especially due to alkaline stress [Citation5]. Alkaline stress deficit dramatically limits the growth and yield for soybean. RNA sequencing is an effective technology to facilitate the understanding of molecular mechanisms in plant stress studies [Citation10]. It has been applied for elucidation of the responses of plants to alkaline stress, including alfalfa and sugar beet [Citation11, Citation12].
However, studies about transcriptome analysis of soybean in response to alkaline stress are limited. Therefore, in this study, we sequenced transcripts from soybean under NaHCO3 stress. Differentially expressed genes were identified and annotated. Functional categorization of responsive genes was conducted to unveil their roles in various pathways and metabolic processes. Furthermore, we focused on the transcription factors and isoflavone related genes and detected their expression changes under alkaline stress by quantitative real-time polymerase chain reaction (qRT-PCR), and suggested the critical roles of these candidate genes under alkaline stress conditions.
Materials and methods
Plant material, growth and tissue harvest
The soybean (DN50) seeds were germinated and grown as reported in a previous study [Citation13]. For alkaline stress, the 12-day-old seedlings were grown in Hoagland’s nutrient solution with 50 mmol/L NaHCO3 for 6 h. The soybean whole seedlings were harvested, and stored at −80 °C for further analysis.
RNA extraction and sequencing of RNA-seq library
Total RNA was extracted from soybean seedlings, using an E.Z.N.A plant RNA kit (Omega Bio-Tek, Beijing), according to the manufacturer’s instructions. RNA samples were sent to BGI-Shenzhen Co. Ltd (Shenzhen, China), and 150-bp paired-end reads were generated using BGI-Seq500 platform [Citation14]. The Wm82.a2.v1 was used as the soybean reference genome.
Sequencing data analysis and identification of differentially expressed genes
After removing adapter and low-quality reads (less than 150 bp) from the raw data, only high-quality tags were retained. All the down-stream analyses performed were based on the high-quality clean data. A reference genome database was constructed and paired-end clean reads were aligned using bowtie2 v2.2.8 and HISAT2v2.0.4. tools [Citation15]. Differentially expressed genes were identified using the Cuffdiff program, and the changes were screened with the threshold |log2ratio| > 1 and FDR < 0.01 [Citation16].
Functional annotation and pathway analysis
The GO functional classification and enrichment analyses were performed by WEGO and agriGO software. The enrichment of KEGG pathway and transcription factors were performed using Benjamini and Hochberg method.
Quantitative real-time PCR
Complimentary DNA (cDNA) was synthesized from total RNA by First Stand cDNA Synthesis kit (Toyobo, Osaka). qRT-PCR was performed using UtraSYBR Mixture (Baioleibo, Beijing) on 7500 Real Time PCR system. The reference gene GmGADPH and gene-specific primers were used to verify the RNA-Seq results of differentially expressed genes (). Relative transcript levels were analyzed using the 2-ΔΔCT method and one-way analysis of variance (ANOVA; *p < 0.05, **p < 0.01) followed by Duncan’s test.
Table 1. Summary of sequencing results.
Results
Quality analysis of the RNA-seq datasets
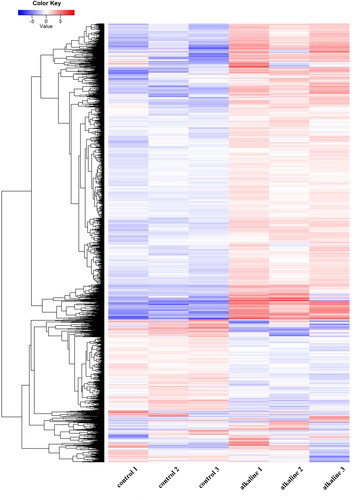
In order to establish the transcriptional profiles of soybean under alkaline stress, total RNA from three samples of plant subjected to alkaline stress were used, while three untreated samples were used for generation of an informative reference transcriptome database. After data filtering, the proportion of the total number of clean read segments was higher, such as the proportions of all clean read segments were over 86.34%, while Q20 values were found greater than 97%. The sequencing sample and the reference genome sequence alignment was higher than 90.39%, and the unique-mapping was over 67.2%. The numbers of clean reads of samples were obtained as shown in .
By calculating the Pearson correlation coefficient of all gene expression amounts between each sample, we identified the correlation coefficient of gene expression between the test samples. The results showed that the correlation coefficient of each sample met the standard, and the gene expression level was similar (). Furthermore, the significantly expressed genes were calculated by FPKM (FPKM > 1). The results showed that 75.7% of the genes were significantly expressed under alkaline stress, compared with 75.0% of the genes in non-treated soybean samples ().
Identification and GO enrichment analysis of differentially expressed genes (DEGs)
In the present study, a total of 6262 genes were identified to have significant changes in their expression level under alkaline stress (|log2 ratio|>1, FDR < 0.01). Among them, 4603 genes were up-regulated and 1659 genes were down-regulated ().
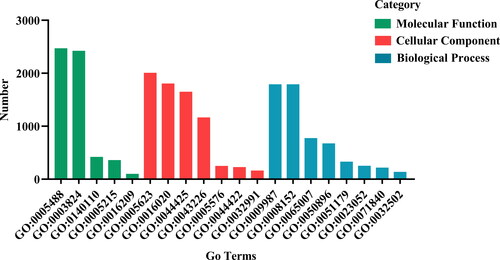
To further investigate the roles of DEGs, we conducted GO enrichment analysis. According to the GO enrichment analysis, the DEGs in alkaline treated soybean were significantly enriched in 43 pathways, including 18 in biological processes, 14 in cell components and 11 in molecular function (). GO analysis also revealed that the DEGs were mainly associated with catalytic activity (GO:0003824 2423 unigenes), binding (GO:0005488 2472 unigenes), metabolic process (GO:0008152 1792 unigenes), cellular process (GO:0009987 1792 unigenes) and cell (GO:0005623 2009 unigenes) in the categories of molecular function, biological process and cellular component ().
Figure 3. Expression pattern analysis of isoflavonoid biosynthesis related genes under alkaline stress. The seedlings were treated with 50 mmol/L NaHCO3 for 6 h. The transcript data results were analyzed using the 2–ΔΔCT method and one-way analysis of variance (ANOVA; *p < 0.05, **p < 0.01) followed by Duncan’s test. The expression levels are normalized relative to GAPDH gene in Glycine max. Data are mean values with standard deviation (±SD).
KEGG pathway analysis and isoflavone involved in the alkaline stress responses
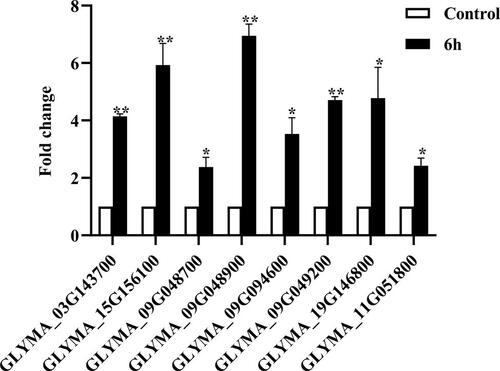
KEGG pathway analysis showed that the DEGs associated with alkaline stress were involved in 133 pathways. For example, most of the DEGs were widely enriched in the pathways of plant–pathogen interaction, MAPK signaling pathway, plant hormone signal transduction, phenylpropanoid biosynthesis, isoflavonoid biosynthesis, flavonol biosynthesis and anthocyanin biosynthesis (Supplemental Table S2). In this study, we observed that the secondary metabolite biosynthesis pathway was the most predominant among pathways, especially the isoflavonoid biosynthesis pathway. To investigate the potential roles of isoflavone in alkaline stress response, we detected the expression patterns of isoflavone biosynthesys related genes using qRT-PCR analysis (). The results showed that eight genes appeared with up-regulated expression levels under alkaline treatment, which are consistent with the results from the transcriptome sequencing data. This finding suggests that isoflavones may play important roles in response to alkaline stress.
Transcription factors involved in the alkaline stress responses
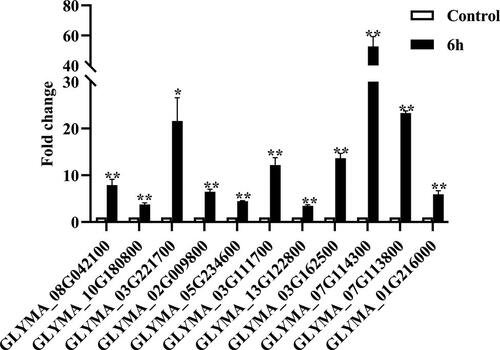
Transcription factors have been found to play a crucial role in response to abiotic stresses, such as salt, drought and cold [Citation17–19]. In this study, many transcription factors were differentially expressed when soybean plants were exposed to alkaline treatment. The comparison showed that the number of DEGs from AP2-EREBF, MYB, WRKY, NAC, bHLH and C2H2 families were comparatively higher than other families (), which suggested that these transcription factors might play significant roles in the response to alkaline stress. Among them, AP2-EREBF and MYB transcription factor families had the largest number of differentially expressed genes. Hence, we focused on AP2-EREBF and MYB-related genes, and 11 AP2-EREBF and MYB genes were selected for further gene expression analysis by using qRT-PCR analysis. Intriguingly, all of these genes appeared with higher transcript levels in response to alkaline stress (). These results are in agreement with the common knowledge that transcription factors might be important for plant in response to alkaline stress.
Figure 4. Differentially expressed genes of transcription factor families under alkaline stress. Differentially expressed transcription factor families were detected under NaHCO3 stress. The x-axis represents the transcription factor families, and the y-axis represents the number of transcription factors. The different colors indicate numbers of transcription factors under alkaline treatment. Blue represents down-regulated DEGs, orange represents up-regulated DEGs.
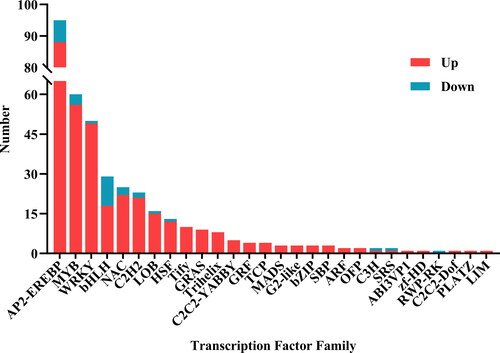
Figure 5. Expression pattern analysis of AP2-EREBF and MYB genes under alkaline stress. The seedlings were treated with 50 mmol/L NaHCO3 for 6 h. The transcript data results were analyzed using the 2–ΔΔCT method and one-way analysis of variance (ANOVA; *p < 0.05, **p < 0.01) followed by Duncan’s test. The expression levels are normalized relative to GAPDH gene in Glycine max. Data are mean values with standard deviation (±SD).
Discussion
Alkaline stress is one of the major detrimental factors for crop productivity [Citation20]. Soybean has been considered one of the most important nutritive value crops [Citation21]. However, the productivity of soybean is significantly limited by alkaline stress. Therefore, to establish the transcriptional profiles of soybean under alkaline stress, we sequenced transcripts from soybean under NaHCO3 stress.
Previous studies have identified a large number of alkali-responsive genes in different plants, such as rice, alfalfa and peach [Citation11, Citation22, Citation23]. This is also supported by our results that the expression levels of a total of 6262 DEGs (including 4603 upregulated and 1659 downregulated) were significantly changed under alkaline stress (). We found that these DEGs were significantly enriched in 43 pathways (), and mainly associated with catalytic activity, binding, metabolic process, cellular process and cell (). These biological processes provide valuable information for understanding the mechanism of soybean response to alkaline stress. For example, the amino acid production and nitrogen metabolism were significantly inhibited in metabolic process under alkaline stress [Citation24]. The catalytic activity and binding process play important roles against alkaline stress in wild jujube [Citation25].
Isoflavonoids are abundantly present in soybean and play multiple roles in plant response to abiotic stresses [Citation26]. Isoflavone metabolism might play a crucial role in response to UV-B stress [Citation27]. Isoflavone synthase genes IFS1 and IFS2 highly correlated with isoflavone accumulation under water deficit conditions [Citation28]. The isoflavone contents were significantly increased in chickpea sprouts germination stage under salt stress [Citation29]. In the present study, we also found some DEGs were enriched in the category of flavonol biosynthesis (Supplemental Table S2). Therefore, we detected the expression patterns of eight isoflavone biosynthesis related genes, and found that these genes were significantly induced under alkaline treatment (). Thus, our results indicated that isoflavones may play important roles in alkaline stress mediated signaling pathways.
Besides isoflavone biosynthesis related genes, we found that some of the DEGs were transcription factors, such as AP2-EREBF, MYB, WRKY, NAC, bHLH and C2H2 families (). This result is consistent with previous studies that these transcription factor families are involved in the response to various abiotic stresses [Citation18]. In this study, the expression of 11 AP2-EREBF and MYB genes were detected by qRT-PCR, and all of them appeared with higher transcript levels under alkaline stress (). The results are in line with the studies that AP2-EREBF and MYB transcription factor families are involved in various abiotic stresses [Citation30, Citation31]. For example, a large number of AP2/ERF genes were induced in response to drought and salinity stresses in durum wheat [Citation32]. GmERF3 was found to be induced by salt, drought, ethylene and ABA, and this category of transcription factors can bind to the GCC box or DRE/CRT element to respond under abiotic stresses [Citation33]. Additionally, MYB transcription factors also play essential roles in response to abiotic stresses in various species, such as in cassava and wheat [Citation34, Citation35]. The ABA-mediated AtMYB96 could improve drought resistance through activation of cuticular wax biosynthesis [Citation36]. Previously, we also isolated an ethylene-responsive gene GsERF71 and found that overexpression of GsERF71 could enhance plant response to alkali stress in Arabidopsis. Similarly, overexpression of soybean GmMYB68 significantly enhanced the resistance to salt and alkali stresses [Citation37]. These findings confirmed that these transcription factors may contribute to the response to alkaline stress, and it might be interesting to study their functional roles and mechanisms in response to alkaline stress.
Conclusions
This study is a comprehensive transcriptome analysis of RNA sequencing in soybean under alkaline stress. Overall, 6262 genes were identified as responsive genes under alkaline stress. The KEGG pathway analysis showed that these responsive genes were involved in 133 pathways. Nineteen AP2-EREBF, MYB and isoflavone related genes were identified, which suggests that these genes can be good candidates for understanding the functions in response to alkaline stress.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability
All data that support the findings reported in this study are available from the corresponding authors upon reasonable request.
Additional information
Funding
References
- Osman KT. 2014. Soil resources and soil degradation. In: Soil degradation, conservation and remediation. Dordrecht: Springer. p. 1–43.
- Yang CW, Jianaer A, Li CY, et al. Comparison of the effects of salt-stress and alkali-stress on photosynthesis and energy storage of an alkali-resistant halophyte Chloris virgata. Photosynt. 2008;46(2):273–278.
- Chaney RL, Coulombe BA, Bell PF, et al. Detailed method to screen dicot cultivars for resistance to fe-chlorosis using fedtpa and bicarbonate in nutrient solutions. J Plant Nutr. 1992;15(10):2063–2083.
- Yang CW, Chong JN, Li CY, et al. Osmotic adjustment and ion balance traits of an alkali resistant halophyte Kochia sieversiana during adaptation to salt and alkali conditions. Plant Soil. 2007;294(1-2):263–276.
- Tuyen DD, Zhang HM, Xu DH. Validation and high-resolution mapping of a major quantitative trait locus for alkaline salt tolerance in soybean using residual heterozygous line. Mol Breeding. 2013;31(1):79–86.
- Karuppanapandian T, Moon JC, Kim C, et al. Reactive oxygen species in plants: their generation, signal transduction, and scavenging mechanisms. Aust J Crop Sci. 2011;5:709–725.
- Toh S, Kamiya Y, Kawakami N, et al. Thermoinhibition uncovers a role for strigolactones in arabidopsis seed germination. Plant Cell Physiol. 2012;53(1):107–117.
- Yu M, Liu ZX, Jiang SS, et al. QTL mapping and candidate gene mining for soybean seed weight per plant. Biotechnol Biotechnological Equip. 2018;32(4):908–914.
- Su DQ, Jiang ST, Wang JJ, et al. Identification of major QTLs associated with agronomical traits and candidate gene mining in soybean. Biotechnol Biotechnological Equip. 2019;33(1):1481–1493.
- Day-Williams AG, Zeggini E. The effect of next-generation sequencing technology on complex trait research. Eur J Clin Invest. 2011;41(5):561–567.
- An YM, Song LL, Liu YR, Shu YJ, et al. De novo transcriptional analysis of alfalfa in response to saline-alkaline stress. Front Plant Sci. 2016;7:931.
- Geng G, Li RR, Stevanato P, et al. Physiological and transcriptome analysis of sugar beet reveals different mechanisms of response to neutral salt and alkaline salt stresses. Front Plant Sci. 2020;11:571864.
- Qiao YH, Lu WX, Wang R, et al. Identification and expression analysis of strigolactone biosynthetic and signaling genes in response to salt and alkaline stresses in soybean (Glycine max). DNA Cell Biol. 2020;39(10):1850–1861.
- Luo D, Zhou Q, Wu YG, et al. Full-length transcript sequencing and comparative transcriptomic analysis to evaluate the contribution of osmotic and ionic stress components towards salinity tolerance in the roots of cultivated alfalfa (Medicago sativa L.). BMC Plant Biol. 2019;19(1):32.
- DuanMu HZ, Wang Y, Bai X, et al. Wild soybean roots depend on specific transcription factors and oxidation reduction related genesin response to alkaline stress. Funct Integr Genomics. 2015;15(6):651–660.
- Yang HH, Zhang YF, Zhen X, et al. Transcriptome sequencing and expression profiling of genes involved in daylily (Hemerocallis citrinaBorani) flower development. Biotechnol Biotechnological Equip. 2020;34(1):542–548.
- Singh K, Foley RC, Oñate-Sánchez L. Transcription factors in plant defense and stress responses. Curr Opin Plant Biol. 2002;5(5):430–436.
- Baillo EH, Kimotho RN, Zhang ZB, et al. Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes. 2019;10:771.
- Khan MS. The role of DREB transcription factors in abiotic stress tolerance of plants. Biotechnol Biotechnological Equip. 2011;25(3):2433–2442.
- Guo R, Yang ZZ, Li F, et al. Comparative metabolic responses and adaptive strategies of wheat (Triticum aestivum) to salt and alkali stress. BMC Plant Biol. 2015;15:170.
- Veiga EV, Vannucchi H, Marchini JS, et al. The nutritive-value of a rice and soybean diet adults. Nutr Res. 1985;5(6):577–583.
- Li N, Liu HL, Sun J, et al. Transcriptome analysis of two contrasting rice cultivars during alkaline stress. Sci Rep. 2018;8(1):9586.
- Sun SX, Song HY, Li J, et al. Comparative transcriptome analysis reveals gene expression differences between two peach cultivars under saline-alkaline stress. Hereditas. 2020;157(1):9.
- Zhang KH, Tang JR, Wang Y, et al. The tolerance to saline-alkaline stress was dependent on the roots in wheat. Physiol Mol Biol Plants. 2020;26(5):947–954.
- Guo MX, Li SP, Tian S, et al. Transcriptome analysis of genes involved in defense against alkaline stress in roots of wild jujube (Ziziphus acidojujuba). Plos One. 2017;12(10):e0185732.
- Zhao MZ, Wang TL, Wu P, et al. Isolation and characterization of GmMYBJ3, an R2R3-MYB transcription factor that affects isoflavonoids biosynthesis in soybean. Plos One. 2017;12(6):e0179990.
- Liu Y, Liu J, Wang Y, et al. The different resistance of two astragalus plants to UV-B stress is tightly associated with the organ-specific isoflavone metabolism. Photochem Photobiol. 2018;94(1):115–125.
- Gutierrez-Gonzalez JJ, Guttikonda SK, Tran LSP, et al. Differential expression of isoflavone biosynthetic genes in soybean during water deficits. Plant Cell Physiol. 2010;51(6):936–948.
- Gao Y, Yao Y, Zhu YY, Ren GX. Isoflavone content and composition in chickpea (Cicer arietinum L.) sprouts germinated under different conditions. J Agric Food Chem. 2015;63(10):2701–2707.
- Yoon Y, Seo DH, Shin H, et al. The role of Stress-Responsive transcription factors in modulating abiotic stress tolerance in plants. Agronomy-Basel. 2020;10(6):788–1041.
- Wang WL, Cui X, Wang YX, et al. Members of R2R3-type MYB transcription factors from subgroups 20 and 22 are involved in abiotic stress response in tea plants. Biotechnol Biotechnological Equip. 2018;32(5):1141–1153.
- Faraji S, Filiz E, Kazemitabar SK, et al. The AP2/ERF gene family in triticum durum: Genome-wide identification and expression analysis under drought and salinity stresses. Genes. 2020;11:1464.
- Zhang GY, Chen M, Li LC, et al. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J Exp Bot. 2009;60(13):3781–3796.
- Ruan MB, Guo X, Wang B, et al. Genome-wide characterization and expression analysis enables identification of abiotic stress-responsive MYB transcription factors in cassava (Manihot esculenta). J Exp Bot. 2017;68(13):3657–3672.
- Wei QH, Chen R, Wei X, et al. Genome-wide identification of R2R3-MYB family in wheat and functional characteristics of the abiotic stress responsive gene TaMYB344. Bmc Genomics. 2020;21(1):792.
- Seo PJ, Lee SB, Suh MC, et al. The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in arabidopsis. Plant Cell. 2011;23(3):1138–1152.
- He YX, Dong YS, Yang XD, Guo DQ, et al. Functional activation of a novel R2R3-MYB protein gene, GmMYB68, confers salt-alkali resistance in soybean (Glycine max L.). Genome. 2020;63(1):13–26.