 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
The quest for discovering novel biomarkers of triple-negative breast cancers (TNBCs) is ongoing. By lacking expression of hormonal receptors, this aggressive subtype of breast cancer presents a clinical dilemma with early recurrence, metastases and poor survival outcome. Filamin-A is a recent novel protein which has proven to play a dual role as an oncogene and a tumour-suppressor in many malignancies. This study analysed the expression of Filamin-A in TNBC cases. Filamin-A was significantly expressed by the majority of the study’s sample, and was correlated with grade, clinical stage and TNM staging. Filamin-A is a versatile protein with numerous interactions with the cytoskeleton components of the tumour cell and with signalling proteins. Its role in modulating chemosensitivity to the chemotherapeutic agent Docetaxel is particularly important, especially in TNBC patients, who have few management options to choose from. This study provides evidence of the clinical significance of Filamin-A in TNBCs, and propose further functional studies to pinpoint the exact function of this protein in this difficult-to-treat subset of breast cancer patients.
Introduction
Worldwide, breast cancer is the most frequently diagnosed life-threatening cancer in women. In most of the developing world, breast cancer remains the most common cause of deaths in women [Citation1]. It is responsible for more than 450,000 global cancer-related mortalities annually worldwide [Citation1,Citation2]. Surgery and radiation therapy, along with adjuvant hormone or chemotherapy when indicated, are now considered primary treatment for breast cancer.
Primary triple-negative breast cancer (TNBC), a sub-type of breast cancer defined by lack of oestrogen receptor (ER), progesterone receptor and human epidermal growth factor receptor 2 and receptor tyrosineprotein kinase erbB2 (ERBB2) gene amplification, represents approximately 15–20% of breast cancer cases [Citation3,Citation4]. TNBC tends to be more aggressive than other subtypes of breast cancer [Citation5]. TNBC cases, as the name implies, lack targeted therapy and despite initial good response to chemotherapy, recurrence of tumour due to development of resistance ensues with development of multiple metastases [Citation6]. All the above makes TNBCs a formidable clinical challenge, with a critical need for the discovery of prognostic biomarkers which could reliably stratify patients with triple-negative disease into different treatment modalities [Citation7].
Filamin-A (FLNA), a cytoskeletal protein with a molecular weight of 280 kDa, crosslinks actin filaments to the orthogonal networks [Citation8]. This protein serves as the scaffold in various signalling networks through its interaction with more than 90 filament-binding proteins [Citation9,Citation10].
As with other membranous and cytoplasmic structural proteins such as the Claudins, Filamins are now known to be critical skeletal supporting proteins. They stabilize the cell through its interaction with actin during cellular movement; but also have far-reaching effects on numerous cellular functions such as cell signalling, cell migration and adhesion, phosphorylation, proteolysis, ion channel regulation, transcription regulation and muscle development [Citation11]. FLNA is one of three members of the Filamins’ family. The level of FLNA is increased in several subtypes of human cancers, including lung, hepatocellular, astrocytic and breast tumours [Citation12–14].
FLNA can function as a modulator of chemosensitivity to docetaxel in TNBC cells through regulation of the MAPK/ERK pathway both in vitro and in vivo [Citation15]. Chemosensitivity to docetaxel and doxorubicin is of prime importance as they are two of the most commonly used chemotherapy agents in breast cancer patients. Knocking-down FLNA, in vitro and in vivo, has made the TNBC tumour cells more sensitive to chemotherapy agents [Citation15]. Tian et al. [Citation16] found that FLNA was associated neuro-vascular invasion and lymph node metastases. In a study of the use of extracellular matrix proteins as serum biomarkers of early-stage breast cancer, a subset of four proteins was found, among which was FLNA, which classified breast cancer patients with 100% sensitivity and 85% specificity [Citation17]. A recent meta-analysis, with weighted gene-expressed network analysis, identified FLNA as one of 11 differentially expressed genes in the protein–protein interaction network with a higher degree (>10) of interactions with other genes and a critical role in the biological processes of the breast cancer cell [Citation18]. FLNA, most recently, was one of ten hub genes dominating several signalling pathways responsible for development and progression of breast cancer, where its knock-down by thioguanate (6-TG) resulted in inhibition of growth of a TNBC cell line [Citation19]. And while expression studies of FLNA in breast cancer has been performed in the past [Citation16,Citation20,Citation21] with variable results ().No study to date has analysed the expression of this versatile protein in TNBCs per se. The aim of this study was to fill this gap.
Table 1. Review of studies analysing the expression of FLNA in breast cancer.
Materials and methods
Ethics statement
The study was approved by the Institutional Review Board (IRB) of the College of Medicine at Alfaisal University.
TNBC tissues
Following approval of the Institutional Review Board (IRB) of the College of Medicine at Alfaisal University, one commercial human microarray (Catalogue no. BR 1509, Biomax, US, Rockville, MD, USA), with 50 cases of triple-negative, invasive ductal breast cancer, triplicate cores per case for a total of 150 TNBC cores; and a second microarray of normal, benign human tissue (Catalogue no.# FDA 992, Biomax US), were used for this study. All cases in the BR 1509 microarray were confirmed triple-negative, invasive ductal breast cancers, which was the only inclusion criterion for this study. Major parameters of these tumours include age, pathological diagnosis, grade, stage and TNM classification.
Immunohistochemical staining
A mouse monoclonal antibody against human Filamin A (10R-F113A; Fitzgerald Industries International, Acton, MA, USA) was diluted 1:300 in antibody diluent (Agilent, Santa Clara, CA, USA) per manufacturer’s instructions; and applied to 5-mm sections from formalin-fixed, paraffin-embedded tissue specimens, using the avidin-biotin peroxidase method (Vectastatin Elite ABC kit; Vector Laboratories, Burlingame, CA, USA), following the manufacturer’s instructions. The IHC stain was performed manually at room temperature. Negative controls were used with omission of primary antibody. A positive control of adrenal tissue (included in the BR 1509 microarray) was used for test optimization and run validation.
Staining evaluation
Filamin-A IHC results
The immunohistochemical staining of FLNA, for all tumours, was evaluated by two pathologists (A.O.) and (E.R.), independently, and a consensus was reached for discordant cases. Because each tumour in the microarray was represented by a triplicate of cores, each core was scored separately then averaged out. All tissues, normal controls, tissues adjacent to cancer or malignant tissue, were compared with the staining of the positive control of adrenal tissue provided in the BR 1509 microarray. The degree of Filamin A reactivity was scored by applying a semi-quantitative immunoreactivity scoring (IRS) system as described by Baccelli et al. [Citation22]. The staining intensity was categorized into four grades: 0, no immunostaining; 1, weak staining; 2, moderate staining and 3, strong staining. The percentage of positive cells was categorized into five grades: 0 (0%); 1, (1–10%); 2 (11–50%); 3 (51–80%) and 4 (>80%). The staining intensity and percentage of positive cells were multiplied to obtain an IRS, in the range 0–12 for each individual case. A case was scored as positive for FLNA with an IRS ≥ 6 defined as positive expression.
Statistical analysis
The χ2 test was used to assess the FLNA expression in TNBCs cases (N = 50). The χ2 test was also used to examine the relationship between FLNA-positive cases (N = 45) and various clinical, demographical and pathological criteria (age, tumour grade, stage and TNM). All statistical analyses were performed with IBM SPSS Statistics software package, version 25.0. Differences were considered statistically significant at the p < 0.05 level.
Results
One copy of the human microarray (Catalogue no# BR 1509, Biomax, US), containing 50 cases of triple-negative, invasive ductal carcinoma, triplicate cores per case; was used for this study. The age of the cancer patients in this array was in the range 33–71, with a median of 50. The TNBCs grades were distributed as follows: 2 patients are grade 1, 26 patients are grade 2 and 22 patients are grade 3. The stages were distributed as follows: 1 patient is stage I, 38 patients are stage II, 11 patients are stage III and 0 patients are stage IV. The Ts (in reference to primary tumour in TNM) were distributed as follows: 2 patients are T1, 34 patients are T2, 11 patients are T3 and 3 patients are T4. The Ns (in reference to the nodal status in TNM) were distributed as follows: 34 patients are N0, 8 patients are N1, 7 patients are N2 and 1 patient is N3. The Ms (in reference to distant metastases in TNM) were distributed as follows: 50 patients are M0, 0 patients are M1.
The designation of TNM is as follows:
T: Primary tumour
T1- Tumour invades submucosaT2- Tumour invades muscularis propriaT3- Tumour invades through muscularis propria into subserosaT4- Tumour directly invades other organs or structures and/or perforate visceral peritoneum
N: Regional lymph nodes
N0- No regional lymph node metastasisN1- Metastases in 1–3 regional lymph nodesN2- Metastases in 4 or more regional lymph nodesN3- Metastases in 10 or more regional lymph nodes, or spread to lymph nodes under the clavicle, or collarbone, or to internal mammary lymph nodesM: Distant metastasesM0- No distant metastasisM1- Distant metastases
The expression of FLNA in normal, benign, unmatched breast tissue (TMA with Catalogue no.# FDA 992, Biomax US); was limited to the myoepithelial cells and basal cell layers, which strongly expressed the protein, whereas the benign epithelial breast cells did not express it (Arrows, ). In TNBC cases, on the other hand, 45/50 cases were positive for FLNA (90% with an IRS of 6–12, χ2=22.967; p < 0.001, ). The rest of the TNBC cases 5 had weak or negative FLNA expression (IRS# 0-5, ). All the FLNA-expressing TNBC cases, 45/45, expressed the protein in the cytoplasm of the tumour cells (Arrows, , ).
Figure 1. FLNA expression (A, B) in normal breast glandular epithelium and ducts. The immune-expression is limited to the myoepithelial and basal cell layers (arrow heads). Normal epithelial cells lack FLNA expression (long arrows). Scale bar for panels A and B = 0.5 mm.
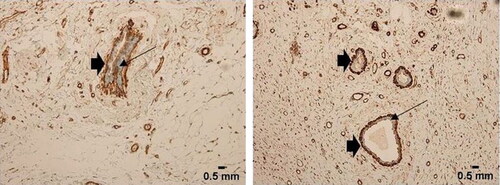
Figure 2. Negative FLNA expression in a few tumours of the study sample. Note the positive endothelial cells and fibrous tissue staining as internal control (Arrows). Tumours (A–C) are all grade 2, stage II, T2N0M0. Scale bar = 2 mm for panels A–C.
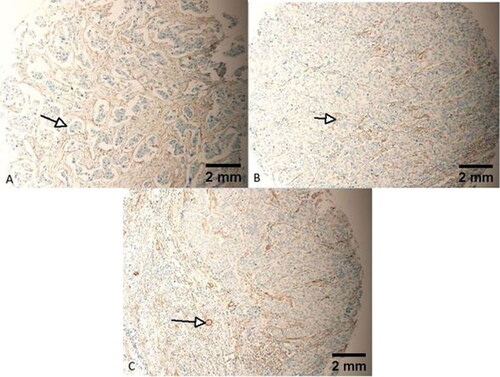
Figure 3. Cytoplasmic expression of all TNBC cells. No nuclear staining was identified. Only cytoplasmic staining of FLNA is identified in TNBC tumour cells in panels A and B (arrows). Scale bar = 2 mm for panels A and B.
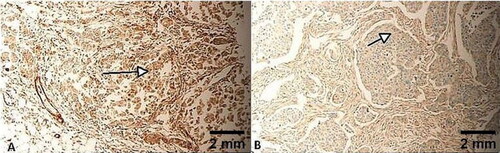
Table 2. FLNA expression and location within the cell.
Association between Filamin-A expression and clinic-pathological parameters of TNBC
The association between Filamin-A expression and clinic-pathological parameters was further analysed in the TNBC tissues (). Positive staining for Filamin-A was significantly correlated with TNBC grade (χ2 = 14.618; p < 0.001); clinical stage (χ2 = 29.783; p < 0.001), T (χ2 = 36.952; p < 0.001) and N (χ2 = 33.217; p < 0.001). There was no significant association between positive staining for Filamin-A and age of the patients (p = 0.394).
Table 3. FLNA-positive TNBC cases clinical, pathological and demographic characteristics.
Morphologically, 35 FLNA-positive tumours (35/45, ) had an IRS score of 12/12 (), 6 FLNA-positive tumours had an IRS of 8 () and 4 of them had an IRS of 6 (). Only 5 TNBCs were negative (). Each of the TNBCs expressed FLNA in the cytoplasm of its cells only, 45/45. No nuclear staining could be identified (). TNBC cells positively expressing FLNA presented with different architectural morphology, including classical, cords-like, spindle and single cells (). One TNBC presented with a tumour embolus with central necrosis, heavily staining with FLNA ().
Figure 4. Heavy cytoplasmic expression of FLNA in all tumour cells, IRS of 12/12. Tumours (A, B) are grade 3, stage II, T3N0M0. Tumour (C) is grade 3, stage III, T4N1M0. Tumour (D) is grade 3, stage III, T4N2M0. Scale bar = 2 mm for panels A–D.
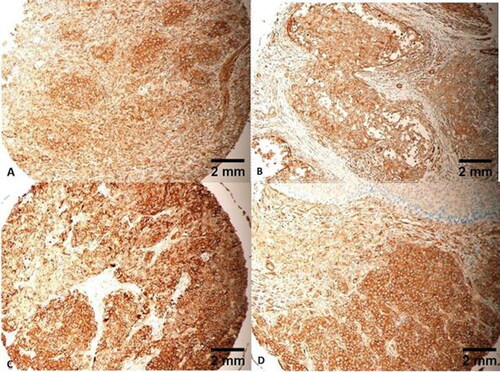
Figure 5. Moderate cytoplasmic expression of FLNA in all tumour cells, IRS of 8/12. Tumours 5 (A–D) are all grade 2 and stage II. Tumours (A, B and D) are T2N0M0. Tumour (C) is T3N0M0. Scale bar = 2 mm for panels A–D.
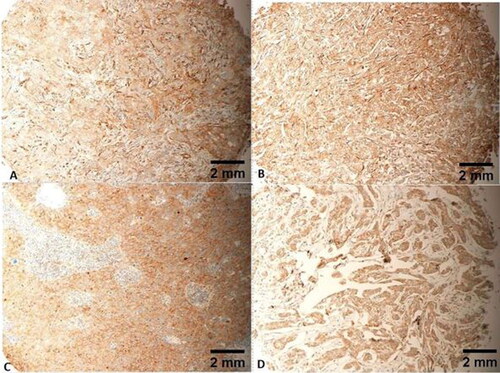
Figure 6. Mild cytoplasmic expression of FLNA in all tumour cells, IRS of 6/12. Tumours (A and C) are both grade 3. Tumours (B and D) are both grade 2. Tumours (A–C) are all stage II. Tumour D is stage I. Tumours (A–C) are all T2N0M0. Tumour D is T1N0M0. Scale bar = 2 mm for panels A–D.
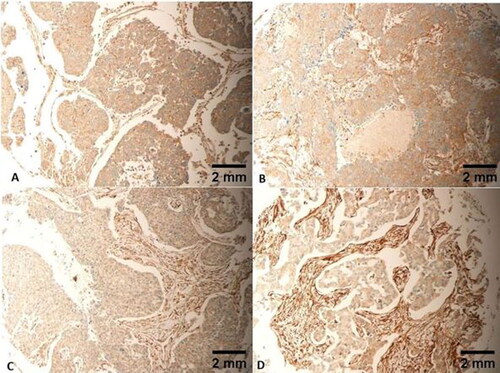
Figure 7. Cytoplasmic expression is noted in TNBCs with different morphology, including classic (A), cords-like (B), spindle (C) and single cell arrangements (D). Scale bar = 2 mm for panels A–D.
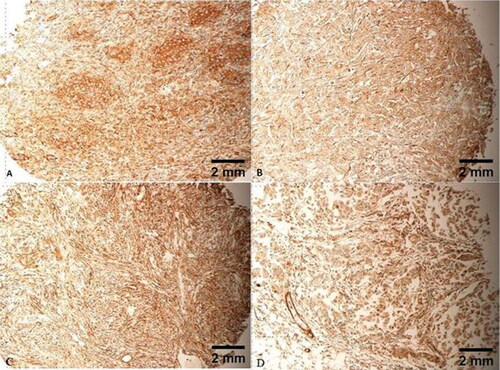
Figure 8. Tumour embolus with strong FLNA expression. Note necrosis at centre of embolus. Scale bar = 0.5 mm.
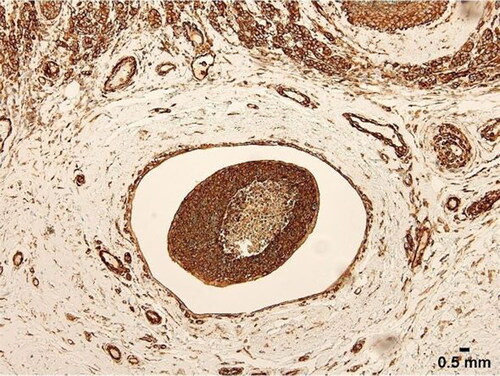
Table 4. Range of IRS scores in FLNA-positive TNBCs.
Discussion
Possible utilization of FLNA in the management of breast cancer stems from its roles in angiogenesis [Citation23], tumour suppression [Citation24], tumour progression [Citation16] modulation of other tumour suppressors such as BRCA1 [Citation25] and modulation of chemotherapeutic agents [Citation26]. The role of Filamin-A in chemotherapy response is agent-dependent as either increasing sensitivity [Citation26] or decreasing it [Citation15]. FLNA-deficient tumour xenografts have been shown to be more sensitive to bleomycin and cisplatin [Citation26]. The fact that Filamin-A, when over-expressed, may decrease the sensitivity of TNBC cells to Docetaxel, is indeed a sobering one, as this chemotherapy agent is on a very short list of management options of TNBCs [Citation15].
This study, which is the first to analyse the prevalence and distribution of FLNA in TNBCs exclusively; and correlate this expression with clinicopathological attributes of these tumours, showed that FLNA was positive in a significant number of the tumours (45/50, 90%, χ2 22.967, p < 0.001). All the FLNA-expressing tumours expressed the protein in their cytoplasm (45/45). It has been recently reported that the cytoplasmic expression of FLNA represents the full length of the protein, its active form and is the most linked to oncogenesis [Citation27]. Proteolysis of the FLNA resulting in its fragmentation and relocation to the nuclei, which results in reduced metastatic potential in cancer [Citation13,Citation21,Citation27]. Interestingly, not a single case in this study exhibited nuclear expression of FLNA. Furthermore, this study’s FLNA expression was significantly linked with the stage, grade and TNM staging (). This is an important finding, given this protein’s active expression in this study’s sample, and may provide a platform for future studies into a possible link between this expression; and the aggressive nature of TNBCs.
Although this is the first such study of FLNA expression in TNBCs, there are other studies that assessed FLNA expression in breast cancer in general. summarizes the findings of those studies. It may be noted from this table that the frequency of FLNA expression varies between the different studies and ranges from 40% to 90%. This could be explained by using different staining techniques, different IHC scoring techniques and the heterogeneous populations of the listed studies. Furthermore, it is noted that the FLNA expression in this study, which stands at 90%, is well above the expression levels of the other studies. TNBCs are known to be aggressive tumours, and FLNA has been shown to be associated with advanced stage breast cancer cases (83% of cases in stage 3 [Citation16]. This may explain the low score of FLNA in the rest of the studies, which could be attributed to the dilutional effects of including other non-TNBC cases in the analysis in those studies. In , it is noted that all of the reported studies agreed on the cytoplasmic expression of the FLNA in breast cancer cells, and the majority of studies also agreed on FLNA expression in normal breast tissue, where the normal breast epithelial cells either has no or very faint expression, whereas the myoepithelial and basal cell layers strongly express this protein. A negative expression of the protein in benign breast tissue is of critical importance, as this will lend further support to the case of utilizing FLNA as a biomarker of management in TNBCs. It could be also noted in that while some studies have shown a dose-like effect when it comes to the expression rates of FLNA in breast cancer-linked to stage [Citation16], which concur with this study’s results, another study of FLNA expressions in breast cancer failed to show such correlations with clinical stage, grade, TNM staging, molecular sub-type and survival studies [Citation20]. To explain this divergent result, it is important to note that the authors of that study [Citation20] have followed an immunohistochemical method which seems to be used for the first time to score the FLNA in their tumour samples, as they have not provided any reference for their method. The authors have used the staining of 10% of tumour cells as their cut-off point, without providing a reason for choosing this threshold, nor providing any reference in that regard [Citation20]. In the opinion of this author, this may have introduced bias, where their group of patients was divided into high and low FLNA based on a randomly-chosen cut-off point, resulting in their non-significant findings. On the other hand, Tian et al. [Citation16] followed the graded method (also used in this study as in [Citation22]) and found significant associations of FLNA expression with grade, score and TNM, albeit with lower FLNA expression at 63.54% [Citation16].
With the numerous interactions of FLNA with components of the cytoskeleton, and other signalling proteins, this creates a complicated picture when discussing its contribution to oncogenesis in general, and in breast cancer particularly. provides a summary of reported mechanisms of FLNA-driven oncogenesis. DNA repair is one proposed mechanism, where FLNA has been proven to be involved in double-stranded breaks repair (DSBR), thus helping tumour cells escape p53 and avoid apoptosis [Citation28]. Moreover, lack of FLNA has been shown to lead to increased susceptibility to DNA damage and cell cycle arrest at the G2/M check point in several cancer types [Citation29]. FLNA provides critical support for tumour cells by promoting angiogenesis, through its close interaction with vascular endothelial growth factor (VEGF) [Citation30]. In breast cancer, knocking-down FLNA expression by RNAi technology resulted in steep knockdown of breast cancer gene 1 (BRCA1) expression [Citation25]. On the other hand, knocking-down FLNA expression in a breast cancer cell line resulted in upregulation of 14-3-3-σ, a postulated tumour-suppressor gene in the breast [Citation31,Citation32]. FLNA interaction with tissue factor (TF) through differential phosphorylation of Ser253 and Ser258 in glandular tumours, including breast cancer, may result in increased surface TF activity, and increased TF’s incorporation and release in microvesicles [Citation33], a process known to promote thrombosis, metastases, tumour growth and tumour angiogenesis [Citation34]. And finally, FLNA’s ability in relaying extracellular matrix signals to the cytoskeleton of the tumour cell, may induce locomotion, through the interaction of FLNA with the actin network [Citation21], integrins [Citation35], migfilin [Citation36] and the Rho family small GTPases [Citation37,Citation38].
Table 5. Proposed mechanisms of the role of FLNA in oncogenesis.
And finally, from a diagnostic point of view, the recent discovery of a novel anti-filamin A antibody which detects a secreted variant of filamin-A in the plasma of patients with breast cancer [Citation39] may provide a much-needed, invaluable, practical test for the diagnostic and follow-up management of TNBC patients.
Conclusions
In summary, Filamin-A may provide a promising biomarker of management in TNBCs. In this study, the normal expression of Filamin-A was limited to myoepithelial and basal-cell layer staining, and the benign epithelial breast cells were negative of the immuno-marker, whereas TNBCs cells had robust and consistent expression by immunohistochemistry. This finding, along with the discovery of a secreted component of filamin-A in the plasma, may aid in the screening, diagnosis and follow-up of TNBCs. A second important aspect of FLNA expression is its potential role in modulating the chemotherapy response in breast cancer patients. The positive expression seen in this study, and the correlations presented, may shed some light on a possible role of this protein in chemotherapy resistance seen in TNBC patients. It may be the first call on specialists in the field to look into the use of this biomarker for the management of this difficult-to-treat subgroup of breast cancer.
Acknowledgement
The author would like to thank Professor Emad Raddaoui, consultant and senior pathologist at King Khaled University Hospital, King Saud University, Riyadh, Saudi Arabia, for his help in the immunohistochemical scoring in this research work.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
The author(s) reported there is no funding associated with the work featured in this article.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019; 69(1):7–34.
- Chen C, Gao D, Huo J, et al. Multiomics analysis reveals CT83 is the most specific gene for triple negative breast cancer and its hypomethylation is oncogenic in breast cancer. Sci Rep. 2021; 11(1):12172.
- Yin L, Duan JJ, Bian XW, et al. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020;22(1):61.
- Swain S. Triple-negative breast cancer: metastatic risk and role of platinum agents. 2008 ASCO Clinical Science Symposium; 2008 June 3; Chicago, IL, USA.
- Blanchard Z, Paul BT, Craft B, et al. BRCA1-IRIS inactivation overcomes paclitaxel resistance in triple negative breast cancers. Breast Cancer Res. 2015;17:5.
- Hudis CA, Gianni L. Triple-negative breast cancer: an unmet medical need. Oncologist. 2011;16(Suppl 1):1–11.
- Wang K, Ash JF, Singer SJ. Filamin, a new high-molecular-weight protein found in smooth muscle and non-muscle cells. Proc Natl Acad Sci USA. 1975;72(11):4483–4486.
- Feng Y, Walsh CA. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat Cell Biol. 2004;6(11):1034–1038.
- Popowicz GM, Schleicher M, Noegel AA, et al. Filamins: promiscuous organizers of the cytoskeleton. Trends Biochem Sci. 2006;31(7):411–419.
- Savoy RM, Ghosh PM. The dual role of filamin a in cancer: can’t live with (too much of) it, can’t live without it. Endocr Relat Cancer. 2013;20(6):R341–56.
- Nallapalli RK, Ibrahim MX, Zhou AX, et al. Targeting filamin a reduces K-RAS-induced lung adenocarcinomas and endothelial response to tumor growth in mice. Mol Cancer. 2012;11:50.
- Ai J, Huang H, Lv X, et al. FlNA and PGK1 are two potential markers for progression in hepatocellular carcinoma. Cell Physiol Biochem. 2011;27(3–4):207–216.
- Lin JF, Xu J, Tian HY, et al. Identification of candidate prostate cancer biomarkers in prostate needle biopsy specimens using proteomic analysis. Int J Cancer. 2007;121(12):2596–2605.
- Zhao P, Ma W, Hu Z, et al. Filamin A (FLNA) modulates chemosensitivity to docetaxel in triple-negative breast cancer through the MAPK/ERK pathway. Tumour Biol. 2016; 37(4):5107–5115.
- Tian HM, Liu XH, Han W, et al. Differential expression of filamin a and its clinical significance in breast cancer. Oncol Lett. 2013; 6(3):681–686.
- Fredolini C, Pathak KV, Paris L, et al. Shotgun proteomics coupled to nanoparticle-based biomarker enrichment reveals a novel panel of extracellular matrix proteins as candidate serum protein biomarkers for early-stage breast cancer detection. Breast Cancer Res. 2020; 22(1):135.
- Cao W, Jiang Y, Ji X, et al. Identification of novel prognostic genes of triple-negative breast cancer using Meta-analysis and weighted gene co-expressed network analysis. Ann Transl Med. 2021;9(3):205.
- Zhang D, An X, Yu H, et al. The regulatory effect of 6-TG on lncRNA-miRNA-mRNA ceRNA network in triple-negative breast cancer cell line. Biosci Rep. 2021 41(2):BSR20203890.
- Joshua LM, Huda F, Rao S, et al. Clinicopathological significance of immunohistochemical expression of filamin a in breast cancer. J Carcinog. 2020;19:13.
- Jiang X, Yue J, Lu H, et al. Inhibition of filamin-A reduces cancer metastatic potential. Int J Biol Sci. 2013;9(1):67–77.
- Baccelli I, Stenzinger A, Vogel V, et al. Co-expression of MET and CD47 is a novel prognosticator for survival of luminal breast cancer patients. Oncotarget. 2014;5(18):8147–8160.
- Li Q, Cao J, He Y, et al. R5, a neutralizing antibody to Robo1, suppresses breast cancer growth and metastasis by inhibiting angiogenesis via down-regulating filamin A. Exp Cell Res. 2020;387(1):111756.
- Xu Y, Bismar T, Su J, et al. Filamin a regulates focal adhesion disassembly and suppresses breast cancer cell migration and invasion. J Exp Med. 2010;207(11):2421–2437.
- Guo Y, Li M, Bai G, et al. Filamin a inhibits tumor progression through regulating BRCA1 expression in human breast cancer. Oncol Lett. 2018;16(5):6261–6266.
- Yue J, Lan S, Yuan C, et al. Prognostic values of Filamin-A status for topoisomerase II poison chemotherapy. Int J Biol Sci. 2012;8(4):442–450.
- Bedolla RG, Wang Y, Asuncion A, et al. Nuclear versus cytoplasmic localization of filamin a in prostate cancer: immunohistochemical correlation with metastases. Clin Cancer Res. 2009;15(3):788–796.
- Yue J, Wang Q, Lu H, et al. The cytoskeleton protein filamin-A is required for an efficient recombinational DNA double strand break repair. Cancer Res. 2009;69(20):7978–7985.
- Meng X, Yuan Y, Maestas A, et al. Recovery from DNA damage-induced G2 arrest requires actin-binding protein filamin-A/actin-binding protein 280 . J Biol Chem. 2004;279(7):6098–6105.
- Uramoto H, Akyurek LM, Hanagiri T. A positive relationship between filamin and VEGF in patients with lung cancer. Anticancer Res. 2010;30:3939–3944.
- Ji ZM, Yang LL, Ni J, et al. Silencing filamin a inhibits the invasion and migration of breast cancer cells by up-regulating 14-3-3σ. Curr Med Sci. 2018 38(3):461–466.
- Phan L, Chou PC, Velazquez-Torres G, et al. The cell cycle regulator 14-3-3σ opposes and reverses cancer metabolic reprogramming. Nat Commun. 2015;6:7530.
- Collier MEW, Ettelaie C, Goult BT, et al. Investigation of the filamin A-dependent mechanisms of tissue factor incorporation into microvesicles. Thromb Haemost. 2017; 117(11):2034–2044.
- Kasthuri RS, Taubman MB, Mackman N. Role of tissue factor in cancer. J Clin Oncol. 2009;27(29):4834–4838.
- Kim H, Sengupta A, Glogauer M, et al. Filamin a regulates cell spreading and survival via beta1 integrins. Exp Cell Res. 2008;314(4):834–846.
- Gehler S, Baldassarre M, Lad Y, et al. Filamin A-beta1 integrin complex tunes epithelial cell response to matrix tension. Mol Biol Cell. 2009;20(14):3224–3238.
- Leung R, Wang Y, Cuddy K, et al. Filamin a regulates monocyte migration through Rho small GTPases during osteoclastogenesis. J Bone Miner Res. 2010;25(5):1077–1091.
- Ohta Y, Hartwig JH, Stossel TP. FilGAP, a rho- and ROCK-regulated GAP for Rac binds filamin a to control actin remodelling. Nat Cell Biol. 2006;8(8):803–814.
- Alper O, Stetler-Stevenson WG, Harris LN, et al. Novel anti-filamin-A antibody detects a secreted variant of filamin-a in plasma from patients with breast carcinoma and high-grade astrocytoma. Cancer Sci. 2009;100(9):1748–1756.
- Ravid D, Chuderland D, Landsman L, et al. Filamin a is a novel caveolin-1-dependent target in IGF-I-stimulated cancer cell migration. Exp Cell Res. 2008;314(15):2762–2773.
