Abstract
The SARS-CoV-2 pandemic is the most devastating health crisis our generation has seen. The present study tries to gather more epidemiological data and improve the knowledge of the demographic factors responsible for a higher incidence of COVID-19. We analyzed the real time polymerase chain reaction (RT-PCR) results obtained in one of the biggest tertiary hospitals in Bulgaria during the first year of the COVID-19 pandemic (May 2020–April 2021). For this period, almost 40% of all tested samples of hospitalized patients and health care workers in University Hospital “St. Marina,” Varna, were SARS-CoV-2 positive. The most affected individuals were 60–79-year-old people. Male sex is a significant risk factor only for the active ages (20–59 years), while both men and women of advanced age have the same risk to be infected with SARS-CoV-2. Interestingly, girls under 19 years were more susceptible to the infection than boys in the same age group.
Introduction
In December 2019, the first cases of new severe pneumonia of unknown origin appeared in Wuhan, China [Citation1]. The infection was later named COVID-19, and the isolated causative agent, SARS-CoV-2. In just a few months, the disease progressed to rapid global distribution and the World Health Organization (WHO) declared COVID-19 to be a pandemic on 11th March 2020.
SARS-CoV-2 belongs to Coronaviridae and retains the general morphology and the life cycle of the family. It has an external lipid envelope with crown-like spikes (S glycoprotein) that facilitates the attachment to the ACE2 receptors and the entry into the host cell. The viral genome consists of single-stranded, non-fragmented RNA with a positive polarity and length of 29.9 kb [Citation2]. The SARS-CoV-2 whole-genome sequencing revealed the Bat-CoV-RaTG13 and the Pangolin-SARS-CoV as the most related animal viruses and possible predecessors to human SARS-CoV-2 [Citation3–5]. Among the known human coronaviruses, SARS-CoV and MERS-CoV are highly similar: 79.5% and 50%, respectively [Citation6, Citation7].
SARS-CoV-2 mainly transmits through the respiratory transmission route via direct contact or aerosols generated from an already infected person [Citation8]. The virus has significant infectious potential and infects all ages, genders, races and social groups. As of December 21, 2021, there have been globally 274,628,461 confirmed cases of COVID-19, including 5,358,978 deaths.
The typical clinical manifestations of COVID-19 include but are not limited to flu-like symptoms, dry cough, high temperature, loss of smell and taste, headache, shortness of breath, pneumonia, diarrhea, dyspnea, acute respiratory distress syndrome and multiple organ failure.
Old age and comorbidities are the main risk factors for symptomatic COVID-19 infection and severe disease [Citation9, Citation10]. Despite this general vision, significant local differences exist in the incidence of SARS-CoV-2: the age structure, behavior and social characteristics of the community have a very complex and often divergent role. The reported inconsistency in the profile of the most vulnerable groups requires gathering data at both global and local levels to draw the entire COVID-19 dynamics picture, which is still unclear.
The present study tries to analyze the frequency of COVID-19 in a university hospital during the first year of the pandemic. We summarized the demographic characteristics of the patients and health care workers who were SARS-CoV-2 positive in the period between 1st May 2020 and 30th April 2021 and discussed the role of age and sex as risk factors for the infection among a hospital’s population.
Subjects and methods
Study area and study design
The present work is a single-center retrospective study of the SARS-CoV-2 testing results obtained between 1st May 2020 and 30th April 2021 in the University Hospital “St. Marina,” Varna, Bulgaria. Varna Administrative Region of Bulgaria is the third-largest region in the country. It is located in the northeastern part of Bulgaria and has 470124 inhabitants (as of 31st Dec 2020). The main health supplier in the region is the University Hospital “St. Marina”, which confronted the highest proportion of the COVID-19 cases, admitting patients from the whole region, as well as from the other neighboring regions. The Virology Laboratory of the hospital has been one of the first accredited laboratories for SARS-CoV-2 diagnostic in Bulgaria. It has been routinely diagnosing SARS-CoV2 suspicious samples since the very beginning of the COVID-19 crisis.
The first positive case in the hospital (and in Varna Region) was detected on 14th March 2020. During the first two months of the pandemic, the detected cases were sporadic and isolated. Therefore, we decided to analyze the frequency of SARS-CoV-2 from the beginning of May 2020. Only data from the standard-of-care clinical laboratory testing were used. No additional sampling or patients’ intervention was performed for this study.
Patients’ selection and sampling
Between 1st May 2020 and 30th April 2021 in the Virology Laboratory of the University Hospital “St. Marina,” a total of 11,076 samples (naso-oropharyngeal swabs and bronchoalveolar lavages) were obtained and examined via multiplex real time polymerase chain reaction (RT-PCR) analysis.
RNA isolation and amplification
RNA extraction was performed with SaMag Viral Nucleic Acid Extraction Kit using a SaMag-12 instrument (Sacace Biotechnologies, Italy) or Maccura Mag-Bind RNA Extraction Kit using an Allsheng Auto-Pure 32 A System (Maccura Biotechnology, China). The initial extraction volumes were 400 µL and 280 µL, and the elution volumes, 50 µL and 30 µL, respectively. Isolated RNAs were amplified with a SARS-CoV-2 Real-TM kit (Sacace Biotechnologies, Italy). This test allows the reverse transcription and amplification to be performed in a single, one-step reaction and detects three genes: a region of the E gene common for all SARS-like coronaviruses (Fam channel), E and N genes specific for SARS-CoV-2 (Rox and Cy5 channels). The sample is considered as SARS-CoV-2 positive if there is an amplification signal with defined Ct value for SARS-like coronaviruses and E- and (or) N-genes of SARS-CoV-2; positive for other coronaviruses if amplification is detected only in the Fam channel and inconclusive if there is an amplification signal for only one of the specific SARS-CoV-2 genes.
Statistical methods
Descriptive statistics – Kruskal-Wallis non-parametric analysis of variance for independent continuous measures and Chi-square test for categorical variables were the preferred statistical methods with a significance level of 0.05 [Citation11]. A logistic regression model was built with the R Project software. Age and sex were included as independent categorical variables (female sex and age < 19 years were encoded as 1) and the SARS-CoV-2 RT-PCR result was the dependent outcome.
Results
Proportion of SARS-CoV-2 positive samples in University Hospital “St. Marina,” Varna, Bulgaria
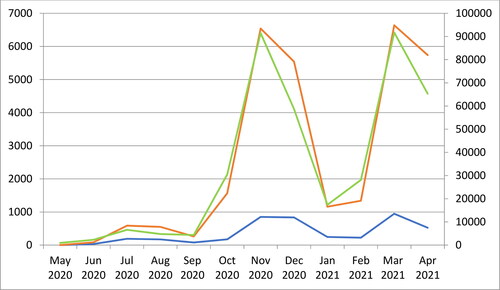
During the first year of the COVID-19 pandemic (from May 2020 to April 2021), we tested for SARS-CoV-2 RNA, a total of 11076 samples of medical staff and patients hospitalized in the University Hospital “St. Marina,” Varna, Bulgaria. Approximately 38.6% of them were positive (4277), 1% (110) were inconclusive samples, and 60.4% (6689) were negative. presents the monthly frequency of the laboratory testing and the proportion of the positive results.
Table 1. Number of positive SARS-CoV-2 cases among hospitalized patients and medical staff of University Hospital “St. Marina,” Varna, Bulgaria.
For the same period, the National COVID-19 Information Portal of Bulgaria (https://coronavirus.bg/) reported a total of 30,013 positive cases in Varna Region and 4,02,187 in Bulgaria. shows that the rates of SARS-CoV-2 positivity in the University Hospital “St. Marina,” Varna, closely followed the overall virus distribution in the country and the region.
Figure 1. Monthly distribution of SARS-CoV-2 positive samples. The blue line represents the number of positive samples in the University Hospital “St. Marina” Varna (left vertical axis); the red line, the positive cases among the total population of Varna Region (left vertical axis) and the green line, all detected COVID-19 cases in Bulgaria (right vertical axis).
Age as a risk factor for SARS-CoV-2 positivity
The median age of the entire cohort was 58 years (range 0–97) and the median age of the positive individuals (63 years) was significantly higher than the median age of those tested negative, 58 years (p < 0.01). As shown in , the risk of testing positive gradually increased with age, and the 60–79 aged individuals were with the highest positivity rate. Interestingly, persons > 80 years had a lower proportion of positive results than the 60–79-years-old people, and the difference between the two groups was statistically significant (Chi-square statistic of 10, p-value = 0.002).
Table 2. Age and gender structure of the positive SARS-CoV-2 samples among hospitalized patients and medical staff of University Hospital “St. Marina,” Varna, Bulgaria.
We calculated the median age of the SARS-CoV-2 positive individuals in the three COVID-19 waves registered between 1st May 2020 and 30th April 2021. We arbitrary divided the first pandemic year into three 4-month periods that generally covered the three waves: from 1st May to 31st Aug 2020; from 1st Sep to 31st Dec 2020; and from 1st Jan to 30th Apr 2021. The median ages of the SARS-CoV-2 infected persons were 55, 64, and 65 years, respectively.
Gender distribution of the SARS-CoV-2 positive individuals
In the University Hospital “St. Marina”, females were slightly more tested with 5684 tests out of 11,076 (51.3%), but males showed significantly higher positivity (40.4% positive samples vs. 36.9% of positive samples among females; Chi-square statistic of 14.9, p-value < 0.001). As seen in , men were of statistically significant higher risk of being positive only in the middle age groups (20-59 years), while in the older age groups there was no correlation between the sex and the PCR positive result: individuals aged > 60 years were of equal risk of becoming infected despite their sex. Moreover, we found that the girls under 19 years were more vulnerable to COVID-19 than boys of the same age and this difference was statistically significant.
Multivariate logistic model of the age and the sex as predictors for infection with SARS-CoV-2
A logistic regression model was run to avoid confounding and to determine the significance of the age and sex as independent risk factors associated with SARS-CoV-2 positivity. Because of the non-linear dependence between the age and the risk of testing positive, age was used as a categorical factor with the same categories as listed in . The results showed that both factors, male sex and advanced age, were independent risk contributors for the tested population ().
Table 3. Multivariate analysis of demographic factors associated with SARS-CoV-2 positivity.
The older age groups had higher odds ratios when compared to the younger patients’ groups: the highest risk group 60–79 years) had an odds ratio of more than six times higher than the control group (<19 years). The odds ratio (male/female sex) was 1.19, and after accounting for sampling variability, the male sex alone could be considered to increase the odds of becoming SARS-CoV-2 infected from 1.10 to 1.28 times when compared to the female sex.
Discussion
This study demonstrates the frequency of SARS-CoV-2 positivity rates among the patients and health care workers in University Hospital “St. Marina,” Varna, Bulgaria. During May 2020 and April 2021, the overall laboratory-confirmed COVID-19 positive results were 38.6% of all tested samples collected from individuals with respiratory infection symptoms or after close contact with an already confirmed case.
In Bulgaria, during the first year of the pandemic, the population’s distribution of COVID-19 formed three distinct peaks: in July–August 2020, in November 2020 and in March 2021 (). The hospital’s dynamic was generally equivalent: it showed the same peaks with some retention in the following weeks. Thus, despite the specificity of the tested population – hospitalized patients and medical staff–the temporal distribution of COVID-19 in the hospital mirrored the epidemic process in the general population.
Age is considered a significant risk factor for hospitalization, severe COVID-19 and death [Citation12, Citation13]. However, the association between age and SARS-CoV-2 infection is not linear. In the general population, the distribution follows a different pattern with the middle ages expressing the highest incidence: the social contacts during working and travelling activities may be in the origin of the higher chance of becoming infected in these age groups [Citation14]. Similarly, in the present study, we showed that the most vulnerable age group for COVID-19 infection was the 60–79-aged individuals, while people over 80 years are less affected. The current study dealt with specific miscellaneous population: on the one hand, tests for non-medical reasons were not performed in our laboratory and the sample consisted mainly of hospitalized patients (i.e. the most severe cases in the general population) but on the other hand, it included not only verified COVID-19 patients but all patients and health care workers that showed respiratory infection symptoms and were suspicious for the infection. We suppose that this is the main reason for the unexpected age profile of the positive individuals in the tested population.
It is a common perception (especially in the mass media) that the age of the affected individuals has been decreasing with the subsequent waves of the pandemic. To test this hypothesis, we calculated the median age of the SARS-CoV-2 positive individuals in the three COVID-19 waves registered between 1st May 2020 and 30th April 2021. Contrary to the general concept, young people in Varna Region did not get more hospitalized with the development of the pandemic and the appearance of the new variants of concern.
The higher risk for men is already well established in the literature [Citation15, Citation16]. However, some studies suggest that women are more vulnerable to COVID-19 [Citation17], or that there is no difference between the sexes [Citation18]. It seems that this depends on the tested population. We suppose that the discrepancy in the reported susceptibility of men versus women comes from the different ages tested in the studies. In the present analysis, men were more susceptible only in the middle age groups (20–59 years). Both sexes in the older age groups were of the same risk. The data obtained allowed us to speculate that females under 19 years were even more vulnerable to COVID-19 than males in the same age group. These results support the well-known hypothesis [Citation19] that the female hormones produced during the active ages and the more healthy lifestyle of women may influence the response to any viral infection, including COVID-19. Despite the statistical significance, the effect of sex is less significant than age and should be analyzed in light of the age findings and other prognostic factors.
Conclusions
This study reveals the importance of demographic characteristics such as age and gender as predictors of positive SARS-CoV-2 results. We confirmed the significance of advanced age and male sex as risk factors for COVID-19 infection among the hospital’s population. However, these factors should be analyzed carefully for each patient as they do not act consistently. Their effect should be always examined in combination with the other well-known predictors for COVID-19 infection, the comorbidities of each patient. Future research should focus to identify rapidly the profile of COVID-19 patients at the greatest risk of adverse outcomes.
Disclosure statement
The authors report no conflict of interest.
Data availability statement
The data that support the findings of this study are available upon reasonable request from the corresponding author. The data are not publicly available due to restrictions, e.g. their containing information that could compromise the privacy of research participants.
Funding
The author(s) reported there is no funding associated with the work featured in this article.
References
- World Health Organization. Novel coronavirus (2019-nCoV) SITUATION REPORT - 1. 2020.
- Khailany RA, Safdar M, Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020;19:100682Jun 1
- Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273.
- Alexandrova R, Beykov P, Vassilev D, et al. The virus that shook the world: questions and answers about SARS-CoV-2 and COVID-19. Biotech. & Biotech. Equip. 2020;35(1):74–102. http://mc.manuscriptcentral.com/tbeq.
- Nie J, Li Q, Zhang L, et al. Functional comparison of SARS-CoV-2 with closely related pangolin and bat coronaviruses. Cell Discov. 2021;7(1):12–21. 6
- Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574.
- Ganesh B, Rajakumar T, Malathi M, et al. Epidemiology and pathobiology of SARS-CoV-2 (COVID-19) in comparison with SARS, MERS: an updated overview of current knowledge and future perspectives. Clin Epidemiol Glob Health. 2021;10:100694.
- Li Q, Guan X, Wu P, et al. Early transmission dynamics in wuhan, China, of novel Coronavirus-Infected Pneumonia. N Engl J Med. 2020;382(13):1199–1207.
- Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069.
- Shahbazi F, Solgi M, Khazaei S. Predisposing risk factors for COVID-19 infection: a case-control study. Casp J Intern Med. 2020;11(Suppl1):495.
- R Development Core Team. The R Project for Statistical Computing [Internet]. [cited 2016 Nov 1]. Available from: https://www.r-project.org/.
- Figliozzi S, Masci P, Ahmadi N, et al. Predictors of adverse prognosis in COVID-19: a systematic review and meta-analysis. Eur J Clin Invest. 2020;50(10):e13362.
- Gupta S, Hayek SS, Wang W, STOP-COVID Investigators, et al. Factors associated with death in critically ill patients with coronavirus disease 2019 in the US. JAMA Intern Med. 2020;180(11):1436–1447. Nov 1
- Salzberger B, Buder F, Lampl B, et al. Epidemiology of SARS-CoV-2. Infection. 2021;49(2):233–239.
- Khan M, Khan H, Khan S, et al. Epidemiological and clinical characteristics of coronavirus disease (COVID-19) cases at a screening clinic during the early outbreak period: a single-Centre study. J Med Microbiol. 2020;69(8):1114–1123.
- Liu R, Han H, Liu F, et al. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin Chim Acta. 2020;505:172–175.
- Karamese M, Ozgur D, Tarhan C, et al. Prevalence of COVID-19 in 10,000 samples from 7853 patients in Eastern Turkey by positive real-time PCR. Future Microbiol. 2021;16(10):697–702.
- Lian J, Jin X, Hao S, et al. Epidemiological, clinical, and virological characteristics of 465 hospitalized cases of coronavirus disease 2019 (COVID-19) from Zhejiang province in China. Influenza Other Respir Viruses. 2020;14(5):564–574.
- Pirhadi R, Talaulikar VS, Onwude J, et al. Could estrogen protect women from COVID-19? J Clin Med Res. 2020;12(10):634–639.
