Abstract
Circular ribonucleic acids (circRNAs) are single-stranded RNAs with covalently closed-loop structures that lack terminal 5′ caps and 3′ poly-(A) tails. Recent evidence confirmed that some circRNAs can be translated into proteins. More importantly, there is a growing body of evidence that dysregulation of circRNAs is closely associated with the development of human diseases, especially malignant tumors. CircZNF609 is a novel circular RNA with an open reading frame and an internal ribosomal entry site that can interact with microRNAs and mRNAs and be translated into proteins. Studies have shown that circZNF609 is abnormally expressed in various human diseases, especially malignant tumors such as hepatocellular carcinoma, nasopharyngeal carcinoma, colorectal cancer and glioma, which lead to the occurrence and development of diseases. This article attempts to provide a comprehensive overview of the structural properties of circZNF609 discovered so far and focuses on the pathogenesis of circZNF609 in human diseases and its potential clinical application value. Finally, we elaborate on the directions of future investigations of this molecule.
Introduction
Unlike linear ribonucleic acids (RNAs), which terminate in 5′ caps and 3′ tails at their ends, circular RNAs (circRNAs) are single-stranded RNAs with covalent closed-loop structures that lack terminal 5′ caps and 3′ poly-(A) tails [Citation1–4]. According to the positions of their parental genes, circRNAs can be divided into exonic circRNAs (ecircRNAs) [Citation5,Citation6], exon-intron circRNAs (EIciRNAs) [Citation7] and intronic circRNAs (ciRNAs) [Citation8,Citation9]. The generation of circRNAs depends on the classical splicing system (splicing site and shear joint), which can be mediated by the reverse complementary elements of flanking introns or by RNA binding protein as well as lariat-mediated formation [Citation10]. Notably, recent reports indicate that circRNAs can also enter extracellular spaces through exosomes, suggesting that they may act as signaling molecules in cell communication, thus demonstrating their immense potential as cancer biomarkers that can be detected using liquid biopsy [Citation11–13].
After circRNA production, most ecircRNAs are exported to the cytoplasm [Citation14,Citation15], whereas ciRNAs and EIciRNAs are mainly found in the nucleus, indicating their potential involvement in transcriptional regulation [Citation16]. CircRNAs in the cytoplasm can serve as molecular sponges that inhibit microRNA (miRNA)-targeted interactions [Citation14, Citation17] and protein functions [Citation18] or as templates for the effective production of peptides through loop-rolling amplifications [Citation19]. In addition, they can act as protein scaffolds to enhance the reaction dynamics of enzyme–substrate interactions. In the cell nucleus, circRNAs modify chromatin and promote gene expression [Citation20]. It is worth noting that some circRNAs that possess internal ribosome entry sites [Citation21–23], such as circMbl [Citation22], circDIDO1 [Citation24] and circZNF609 [Citation25], can be translated into proteins in a cap-independent manner using the start codons.
The structure of circRNAs is highly stable. Researchers have found that linear RNAs can be degraded by RNase R; however, circRNAs are naturally resistant to RNase R [Citation26,Citation27]. The evolution of circRNAs is known to be highly conserved, based on three-generation nanopore sequencing and circRNA identifier using long-read sequencing data (CIRI-long) analysis, which provides direct full-length evidence of circRNAs using nanopore sequencing [Citation28]. The sequencing results demonstrated that consistent full-length sequences of circRNAs could be obtained with higher accuracy, which is of great help in circRNA research [Citation29]. In recent years, circRNAs have attracted great interest because of their important roles in the onset and progression of human diseases, especially in tumorigenesis [Citation30–35]. Therefore, deeper insight into how circRNAs are implicated in stemness, drug resistance and their potential role as biomarkers of cancer may help advance the clinical strategies for effective tumor treatment [Citation30, Citation36].
The circRNA zinc-finger protein 609 (circZNF609) has been confirmed to exert a significant role in human diseases. CircZNF609, as a newly discovered molecule, exerts a vital influence on human diseases [Citation37–39], especially in cancers such as breast cancer [Citation40], gastric cancer (GC) [Citation41,Citation42] and glioma [Citation43,Citation44]. Importantly, circZNF609 expression was proven to be associated with the progression and prognosis of many human diseases, especially cancers. In this review, the structural properties and functions of circZNF609 are summarized, and its possible clinical applications are also explored. In this review, we made an attempt to collect all the published articles on circZNF609 and human diseases, providing a comprehensive description of the currently known role of circZNF609 in human diseases, and we will also discuss the future research directions of circZNF609.
Biological features of circZNF609
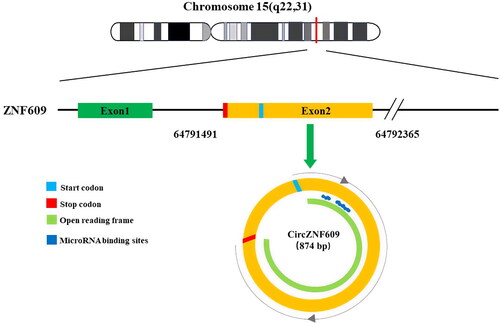
CircZNF609 is a circRNA produced from exon 2 of the zinc-finger protein 609 (ZNF609) gene. The structure of this circRNA was revealed by genomic sequence [Citation45]. CircZNF609 was initially found to be expressed in growing myoblasts, and the proliferation of myoblasts was inhibited when circZNF609 was knocked out [Citation25]. CircZNF609, whose circBase ID is has_circ_0000615, is located at chr15: 64791491–64792365 and comprises 874 base pairs (). CircZNF609 is formed by non-linear splicing of the ZNF609 pre-mRNA [Citation25]. Similar to other circRNAs, circZNF609 is a single-stranded circRNA without 5′ caps and 3′ tails at its ends. Additionally, it has been confirmed through northern blot analysis that circZNF609 was susceptible to si-circZNF609 treatment and resistant to RNase R, while the linear ZNF609 mRNA was resistant to si-circZNF609 treatment and susceptible to RNase R [Citation25, Citation46]. Based on bioinformatics, the CSCD Database (whu.edu.cn) indicates that circZNF609 binds to miRNA at 9,606 sites in the human species. Reportedly circZNF609 shows stable expression in various human tissues, and is preferentially located in the cytoplasm [Citation43, Citation47].
Figure 1. Generation of circZNF609. CircZNF609 is derived from exon 2 of ZNF609. Its circBase ID is hsa_circ_0000615. CircZNF609 is located at chr15: 64791491–64792365 and comprises 874 base pairs. circZNF609 was predicted to have 12 open reading frames (ORFs) in the NCBI database, and 9606 microRNA binding sites in human species were found in the CDSD database.
Roles of circZNF609
As a therapeutic target
More and more studies are exploring the possibility of applying circRNAs as therapeutic agents and molecular targets [Citation48,Citation49]. At the same time, with the in-depth research focusing on the sponge ability of circZNF609 molecules, more and more evidence indicates that circZNF609 has the potential to become a clinical therapeutic target [Citation50]. In Hepatocellular Carcinoma (HCC), circZNF609 enhanced cancer cell proliferation, invasion and is associated with poor overall survival, indicating that circZNF609 might be a promising target in HCC [Citation47]. In non-small cell lung cancer (NSCLC), high expression of circZNF609 was associated with lymph node metastasis and an advanced clinical stage of NSCLC [Citation51].
Interact with mRNA
The latest studies found that there is a biological function of the interaction between circRNA and mRNA based on circZNF609 [Citation52,Citation53]. Rossi et al. [Citation53] found that the circZNF609 sequence across the back-splicing junction (BSJ) interacted with Cytoskeleton Associated Protein 5 (CKAP5) mRNA and regulated CKAP5 protein levels via binding ELAV-like protein 1 (ELAVL1). CKAP5 protein has a significant role in mitotic progression [Citation54,Citation55]. The depletion of circZNF609, and hence the downregulation of CKAP5 protein levels through CKAP5 mRNA-circZNF609 interaction, lead to the dysregulation of microtube dynamics, abnormal mitotic progression and chromosome segregation, and accumulation of DNA damage. What is more, the knockdown of circZNF609 could also strengthen the anti-tumor effects of microtubule (MT)-targeting drugs [Citation52,Citation53]. The CKAP5 mRNA-circZNF609 interaction might operate in all tumor cells that overexpress circZNF609, because the interaction occurred at the level of the BSJ, which is unique to the circular RNA. And the data demonstrated that the use of siRNAs against circZNF609 or locked nucleic acid (LNA)-modified oligonucleotides preventing CKAP5 mRNA-circZNF609 interaction might be adjunctive therapeutic strategies to prevent tumor growth [Citation53].
Encode a protein
Interestingly, 12 open reading frames (ORF) were predicted from the circZNF609 sequence using the ORF-Finder (NCBI) database (https://www.ncbi.nlm.nih.gov/orffinder/) in total. Researchers have found that circZNF609 contains a 753-nt ORF, which includes the transcription start sites and stop codons, and can undergo translation in a cap-independent manner (). In addition, the untranslated region (UTR) of circZNF609 acts as an internal ribosome entry site (IRES) in a splice-dependent manner to facilitate circRNA translation by recruiting ribosomes [Citation25, Citation56]. Moreover, this translation is induced by heat stress at a temperature of 44 °C for 3 h [Citation25]. It was found that circRNA was translated at a lower efficiency compared with the linear form [Citation25]. Studies have demonstrated that circZNF609 expression is influenced by N6-methyladenosine modification, and both YTH N6-Methyladenosine RNA Binding Protein 3 (YTHDF3) and eukaryotic translation initiation factor 4 gamma 2 (eIF4G2) proteins are physically associated with the endogenous circZNF609. Splicing plays a crucial role in the production of circZNF609 proteins because splice site deletion results in the complete loss of these proteins [Citation56,Citation57]. However, the function of the protein translated by circZNF609 must be studied further.
circZNF609 in tumors
CircZNF609 in the nervous system
Glioma
Glioma is a type of tumor that occurs in the brain and spinal cord. It is the most prevalent primary intracranial tumor [Citation58,Citation59].
CircZNF609 has been identified as an oncogenic circRNA in gliomas. It could promote glioma cell migration and proliferation by acting as a ceRNA for miR-134-5p (, ) [Citation44]. Compared with normal cells, the expression of circZNF609 was significantly increased in three glioma cell lines. They found that circZNF609 served as a ceRNA and regulated the miR-134-5p/BTG-2 signaling pathway to promote glioma cell migration and proliferation [Citation44]. Recently, Du et al. [Citation43] showed that the expression of circZNF609 in high-grade glioma tissues was higher than that in low-grade glioma tissues. Moreover, overexpression of circZNF609 could negatively regulate miR-1224-3p to increase Polo-like kinase 1 (PLK1) expression, leading to cell proliferation and migration. Zhao et al. [Citation60] discovered that the depletion of circZNF609 repressed cell viabilities, proliferation, invasion, glycolysis, and stimulated cell apoptosis. Meanwhile, the depletion of circZNF609 suppressed cell growth in nude mice. Therefore, researchers concluded that circZNF609 contributed to cell survival and glycolysis via targeting miR-378/SLC2A1 axis. These results present the new function of circZNF609 in regulating glioma and provide new evidence of the role of circular RNAs in glioma development [Citation60].
Figure 2. Functional roles of circZNF609 acting as microRNA sponges in human cancers. The figures of human organs are from a free medical figure supplier Servier Medical Art (smart.servier.com).
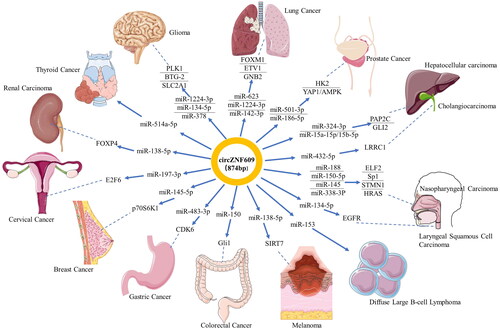
Table 1. Functional characteristics of circZNF609 in multiple human cancers.
CircZNF609 in the respiratory system
Lung cancer
Lung cancer is classified into two major types based on the appearance of lung cancer cells under the microscope: small cell lung cancer (SCLC) and non-SCLC (NSCLC). Approximately 85% of lung cancers are NSCLC which includes lung adenocarcinoma (LUAD) [Citation77].
Wang et al. [Citation51] reported that circZNF609 was upregulated in NSCLC tissues compared to corresponding noncancerous samples. Moreover, circZNF609 expression in NSCLC negatively correlated with patients’ survival. High circZNF609 expression was associated with lymph node metastasis and an advanced clinical stage of NSCLC. In vivo and in vitro experiments demonstrated that circZNF609 promoted the progression of NSCLC by sponging miRNA-623 and upregulating the expression of forkhead box protein M1 (FOXM1) [Citation51]. Zuo et al. [Citation61] revealed that circZNF609 participated in the development and progression of LUAD. The study demonstrated that circZNF609 promoted ETS variant transcription factor 1 (ETV1) expression by sponging miR-1224-3p in LUAD cells [Citation61]. In addition, Liu et al. [Citation50] found that G protein subunit beta 2 (GNB2) was a downstream target of the circZNF609/miR-142-3p axis, promoting cell proliferation and migration of lung cancer cells. Overall, this accumulating evidence suggests that circZNF609 plays an oncogenic role in lung cancer and may be a useful prognostic biomarker and therapeutic target.
Laryngeal squamous cell carcinoma
Laryngeal squamous cell carcinoma (LSCC) accounts for approximately 90% of all malignancies in the larynx and is the second most common malignancy of the respiratory system [Citation78].
Yin et al. [Citation62] initially reported that circZNF609 was significantly upregulated in LSCC and associated with poor survival of LSCC patients. The study suggested that circZNF609 could present tumor-promoting effect to promote LSCC invasion, proliferation and metastasis by regulating miR-134-5p, thus activating the Epidermal growth factor receptor (EGFR), illustrating that circZNF609 might be an oncogene in LSCC [Citation62]. The finding would provide a new perspective for the study of LSCC.
Nasopharyngeal carcinoma
Nasopharyngeal carcinoma (NPC) is caused by the abnormal growth of nasopharyngeal epithelial cells and is highly associated with Epstein–Barr virus infection and is characterized by low differentiation and early metastasis [Citation79,Citation80].
The function of circZNF609 in NPC was first investigated by Zhu et al. [Citation64]. CircZNF609 expression was elevated in NPC cell lines compared to normal nasopharyngeal epithelial cell line NP69. CircZNF609 overexpression promoted NPC progression by binding to miR-150-5p, resulting in the elevated expression of the specificity protein 1 (Sp1) [Citation64]. Li et al. [Citation63] found that knockdown of circZNF609 in NPC cells suppressed cell cycle transition from the G0/G1 stage to the S stage and increased cell apoptosis. Furthermore, circZNF609 overexpression decreased miR-188 levels and increased E74-like ETS transcription factor 2 (ELF2) levels, leading to the progression of NPC. Moreover, CircZNF609 promoted the multiplication, migration and angiogenesis of NPC cells by regulating the miR-145/STMN1 axis [Citation46]. In addition, circZNF609 was found to be a competing endogenous RNA (ceRNA) to regulate the expression of the Harvey rat sarcoma virus oncogene homolog (HRAS) by inhibiting miR-338-3p to promote the proliferation, migration, invasion and glycolysis of NPC cell. In vivo, depletion of circZNF609 inhibited NPC tumor growth [Citation65]. In summary, these studies show that circZNF609 might play an oncogenic role in NPC and could be a potential prognostic indicator and therapeutic target for NPC.
CircZNF609 in the haemopoietic system
Diffuse large B-cell lymphoma
Diffuse large B-cell lymphoma (DLBCL) is one of the most common subtypes of Non-Hodgkin Lymphoma (NHL), which is a common malignant tumor originating from the lymphohematopoietic system [Citation81]. DLBCL has a rapid growth rate and a high degree of malignancy. Thus, it is indispensable to find effective methods to inhibit the development of DLBCL.
Yang et al. [Citation66] reported that circZNF609 was highly expressed in DLBCL, and miR-153 expression was down-regulated in DLBCL [Citation66]. The research showed that circZNF609 could inhibit the proliferation of DLBCL OCI-LY19 cells, block cell cycle progression and induce cell apoptosis by targeting miR-153 regulation. Consequently, circZNF609 could be regarded as a potential target for targeted gene therapy for DLBCL.
CircZNF609 in the digestive system
Hepatocellular carcinoma
HCC, the most common primary liver cancer, typically occurs in the context of chronic liver disease, is highly invasive and has a poor prognosis [Citation82,Citation83].
CircZNF609 executes its oncogenic functions in HCC, as described by Liao et al. [Citation67]. CircZNF609 was upregulated in HCC cell lines compared to normal liver cells. They found that circZNF609 could accelerate the progression of HCC by regulating the miR-324-3p/PAP2C regulatory network. Moreover, circZNF609 overexpression in HCC cells was associated with a poor prognosis of HCC. Studies have shown that circZNF609 might mediate the activation of the Hedgehog pathway to increase cell metastasis-associated proteins, EMT-related proteins and stemness-linked proteins by inhibiting miR-15a-15p/15b-5p expression and elevating GLI Family Zinc Finger 2 L (GLI2) expression [Citation47]. These results consistently indicate that circZNF609 contributes to the development of HCC, which may provide a new clue for researchers in their exploration of better treatments for HCC.
Gastric cancer
Although its occurrence has declined significantly over the past two decades, GC is still among the most prevalent cancers worldwide [Citation77, Citation84].
Wu et al. [Citation42] observed that circZNF609 expression positively correlated with a more advanced tumor stage in GC patients, suggesting the potential oncogenic effect of circZNF609 on GC development. The proliferation and migration of GC cells were markedly accelerated by circZNF609 overexpression, which negatively correlated with the 5-year survival rate of GC. The finding indicated that the circZNF609/miR-483-3p/CDK6 axis is potentially involved in GC development [Citation42]. In addition, circZNF609 has been shown to promote the proliferation and invasion of GC cells and inhibit their apoptosis [Citation41]. Further analysis indicated the diagnostic value of circZNF609 in GC [Citation42]. These findings show that the upregulation of circZNF609 denotes poor prognosis in GC patients and may serve as a disease marker.
Colorectal cancer
Colorectal cancer (CRC) is a heterogeneous disease that originates from the intestinal epithelium. In CRC there is typically accumulation of mutations and dysregulation of the immune responses [Citation85,Citation86]. This remains one of the most commonly diagnosed cancers to date [Citation77].
Wu et al. [Citation68] firstly explored the relationship between circZNF609 and CRC. They found that circZNF609 could arrest the cell cycle in the G0/G1 phase. Mechanistically, they found that circZNF609 could absorb miR-150 to promote the expression of GLI Family Zinc Finger 1 (Gli1), thereby promoting the proliferation, migration, and invasion of CRC cells, indicating that circZNF609 acted as a tumor-promoting factor in CRC [Citation68]. CircZNF609 depletion upon doxycycline induction decreased the proliferation of HCT cell lines. In a tumor xenograft model, circZNF609 overexpression in cancer cells increased tumor growth. Therefore, circZNF609 is thought to accelerate CRC progression [Citation87].
However, Zhang et al. [Citation69] found that circZNF609 was downregulated in the serum of CRC patients, leading to cancer cell apoptosis and inhibition of cell proliferation via p53 upregulation. The circZNF609 levels in serum samples from CRC patients were significantly lower compared to healthy controls. CircZNF609 overexpression in HCT116 cells had little effect on the expression of proliferating cell nuclear antigen (PCNA) and cellular Myc (c-Myc), which were indicators of cell proliferation activity. Moreover, Bax and p53 were upregulated, while Bcl-2 expression was downregulated in the circZNF609 overexpression group [Citation69].
The conflicting results in different reports suggest that circZNF609 plays different roles in CRC under various conditions.
Cholangiocarcinoma
Cholangiocarcinoma (CCA), the second most common hepatic malignancy after HCC [Citation88], is a highly lethal adenocarcinoma of the hepatobiliary system and can be classified as intrahepatic, irritative and distal.
It was found that circZNF609 was highly expressed in CCA, and high expression of circZNF609 was significantly related to advanced TNM stage, lymphatic invasion and survival time [Citation70]. In the study, circZNF609 was found to promote the proliferation, migration and invasion of CCA cells in vitro and the tumor growth in vivo. CircZNF609 plays an oncogenic role in CCA by upregulating Leucine Rich Repeat Containing 1 (LRRC1) expression by sponging miR-432-5p. Additionally, researchers noticed that the YY1 Transcription Factor (YY1) could bind to the promoter of ZNF609 to promote circZNF609 transcription. RNA binding protein elF4A3 (Eukaryotic Translation Initiation Factor 4A3) promoted the circZNF609 circular formation by combining with pre-mRNA of circZNF609. All in all, YY1/elF4A3/circZNF609/miR-432-5p/LRRC1 participates in the progression of CCA, and circZNF609 is expected to be a novel biomarker for targeted therapy and prognostic evaluation of CCA [Citation70].
CircZNF609 in the urinary system
Renal carcinoma
Renal carcinoma is a well-known tumor affecting the urinary system [Citation89,Citation90]. Although the survival rate of renal carcinoma has improved, metastasis remains a major challenge in treating this cancer [Citation91].
Xiong et al. [Citation71] found that circZNF609 might have clinical implications for the diagnosis and treatment of renal carcinoma. CircZNF609 expression in renal carcinoma cell lines was higher than that in the renal epithelial cell line, indicating that circZNF609 might initiate renal carcinoma. CircZNF609 regulated forkhead box P4 (FOXP4) expression by binding to miR-138-5p to accelerate renal carcinoma progression [Citation71]. These studies suggest that circZNF609 may be a potential target for renal carcinoma therapy.
Prostate cancer
Prostate cancer (PCa) remains the second leading cause of cancer-related deaths in men worldwide [Citation92].
CircZNF609 is potentially a novel therapeutic target for PCa [Citation72,Citation73]. Jin et al. [Citation72] showed that circZNF609 was highly expressed in PCa tissues. CircZNF609 overexpression downregulated miR-186-5p to activate the YAP1 and AMPK signaling pathways and promote the multiplication, migration and invasion of PCa cells, and inhibited PCa cell apoptosis [Citation93,Citation94]. Thus, circZNF609 can influence the progression of PCa cells by modulating miR-186-5p.
Aerobic glycolysis, a hallmark of cancer, is advantageous for the proliferation of cancer cells [Citation95]. Du et al. [Citation73] found that the survival fraction of PCa cells, glucose uptake and lactate production were markedly reduced by the interference of circZNF609. Radioresistance is an obstacle to PCa treatment [Citation96,Citation97] because it was elevated in vitro and in vivo after circZNF609 overexpression in PCa cells. The circZNF609/miR-501-3p/HK2 axis promotes migration, invasion, glycolysis and radioresistance, and inhibits the apoptosis of PC3 and LNCaP cells [Citation73]. These results indicate that circZNF609 might promote PCa progression, making it a potential therapeutic target and prognostic biomarker in PCa.
CircZNF609 in the reproductive system
Breast cancer
Breast cancer is one of the most widespread cancers among women worldwide [Citation98]. Wang et al. [Citation40] showed that circZNF609 expression was increased in breast cancer tissues and cell lines. CircZNF609 expression was prominently related to lymph node metastasis, Ki-67 (a marker of cell proliferation) level, advanced TNM stage and poor overall survival. Additionally, circZNF609 could act as a sponge of miR-145-5p to promote ribosomal protein S6 kinase β1 (p70S6K1) expression, thereby promoting the proliferation, migration, and invasion of breast cancer cells [Citation40]. Overexpression of circZNF609 predicts a poor prognosis in breast cancer.
Cervical cancer
Cervical cancer (CC) is among the most pervasive cancers in women [Citation77, Citation99] and occurs mostly in women aged > 30 years. Prolonged infection with certain types of human papillomavirus is the main cause of CC [Citation100].
CircZNF609 was found to be vital to CC progression, suggesting that it could be used as a therapeutic target [Citation74]. The expression of circZNF609 was markedly higher in CC tissues than in normal human endocervical epithelial cell lines. In addition, E2F transcription factor 6 (E2F6) is a downstream target through which circZNF609/miR-197-3p promotes CC cell proliferation, migration and invasion in vitro [Citation74]. Therefore, circZNF609 might be used as a predictor and a therapeutic target of CC.
CircZNF609 in endocrine system
Thyroid cancer
Thyroid cancer (TC) stands out as one of the most predominant endocrine malignancies [Citation101,Citation102]. Advances have been made in recent years in the treatment of thyroid cancer, including targeted biological therapies and adjunctive radiotherapy, but some TC patients suffer treatment failure, resulting in local recurrence and distant metastases [Citation103].
Shi et al. [Citation75] found that circZNF609 functions as an oncogene in TC development via downregulating miR-514a-5p, promoting TC cells proliferation, invasion and migration in vitro and accelerating tumor growth in vivo. This provides a new perspective for understanding and implementing molecular targeted therapy of TC.
CircZNF609 in other systems
Rhabdomyosarcoma
Rhabdomyosarcoma (RMS) is one of the most predominant types of soft tissue sarcomas in children. It originates from mesenchymal progenies and is a high-grade tumor of skeletal myoblast-like cells with a high incidence of metastasis and poor prognosis [Citation104–106]. In children, RMS is usually classified into two major histological subtypes: embryonal rhabdomyosarcoma (ERMS) and alveolar rhabdomyosarcoma (ARMS). ARMS is characterized by a lack of sensitivity to treatment and poor prognosis [Citation106,Citation107].
Rossi et al. [Citation45] found that circZNF609 was upregulated in RMS cell lines, which helped sustain the aberrant proliferation of cancer cells. Depletion of circZNF609 affected the cell cycle and triggered the expression of immune response genes. The results revealed that circZNF609 depletion caused ERMS inhibition in vivo by reducing the p-Rb/Rb ratio and p-Akt protein levels to induce G1/S arrest. However, the depletion of circZNF609 did not affect the growth of ARMS cell lines, suggesting that circZNF609 alone may not be enough to affect cell growth of ARMS [Citation45]. Overall, circZNF609 is upregulated in RMS and might be an effective therapeutic target for ERMS.
Melanoma
Melanoma, the most prevalent skin cancer, has a poor prognosis and survival rates. Moreover, the incidence of melanoma has rapidly increased [Citation108–110].
Liu et al. [Citation76] found that depletion of circZNF609 significantly reduced the viability, invasion and migration and induced apoptosis of melanoma cell lines. The comet assay, which is used to measure DNA damage in individual cells [Citation111], revealed that DNA damage was enhanced by circZNF609 depletion in melanoma cells, while the phenotype could be blocked by miR-138-5p or Sirtuin 7(SIRT7) overexpression. Therefore, circZNF609 could suppress DNA damage by modulating the miR-138-5p/SIRT7 axis [Citation76]. Altogether, these studies suggest that circZNF609 might function as a tumor promoter in melanoma [Citation76].
Clinical features of circZNF609 in tumors
As mentioned before, circZNF609, like other circRNAs, blocks circulation and resists degradation by exonuclease, thus maintaining an extended and stable existence in cells [Citation26]. Because of its stability, it has the potential to be a valuable biomarker for clinical diagnosis. Additionally, circZNF609, as a newly discovered molecule, has an internal ribosome entry site that uses an initiation codon to translate in a cap-independent manner [Citation25].
Through bioinformatics, statistics and research on various types of cancer patients, we confirmed that circZNF609 is closely related to a variety of clinical features of patients (). CircZNF609 expression, which acts as an oncogenic circRNA, is significantly elevated in various cancers. In vivo, circZNF609 overexpression promoted tumor growth in tumor xenograft models. Lymph node metastasis and distant metastasis are usually characterized by poor prognosis, which can be observed in cancer patients with high expression of circZNF609, such as in patients with lung cancer [Citation50,Citation51, Citation61], NPC and other cancers [Citation46, Citation63–65]. In addition, the abnormal expression of circZNF609 was associated with low overall survival and advanced TNM stages. Du et al. [Citation73] also noted that circZNF609 is associated with radiosensitivity, ultimately affecting PCa treatment. These findings suggest that circZNF609 is a promising target for cancer therapy.
Table 2. Clinical features associated with circZNF609 in multiple human cancers.
circZNF609 in non-malignant diseases
Hirschsprung’s disease
Hirschsprung’s disease (HSCR) is characterized by an absence of ganglion cells in the distal bowel, beginning at the internal sphincter and extending proximally. The resulting aganglionic segment of the colon fails to relax, causing a functional obstruction [Citation112]. Therefore, it is necessary to study the pathogenesis of HSCR and develop new treatment methods.
Peng et al. [Citation113] found that circZNF609 and miR-150-5p (, ) may participate in the pathogenesis of HSCR. It was shown that circZNF609 was significantly downregulated in HSCR compared with bowel tissues. The finding revealed that the proliferation and migration of cells was attenuated dramatically after circZNF609 interference. The research illustrated that circZNF609 took part in the onset of HSCR through the crosstalk with AKT Serine/Threonine Kinase 3 (AKT3) by competing for shared miR-150-5p [Citation113]. Thus, it provides a novel insight to diagnose the HSCR and develop new treatment methods.
Figure 3. Functional roles of circZNF609 acting as microRNA sponges in human non-malignant diseases. The figures of human organs are from a free medical figure supplier Servier Medical Art (smart.servier.com).
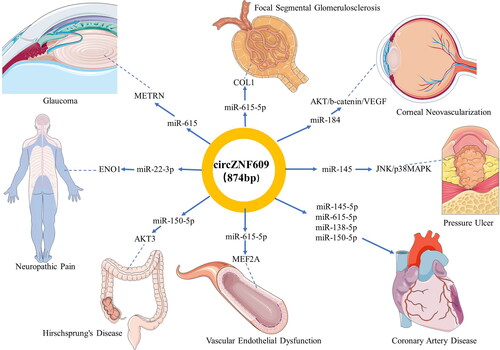
Table 3. Functional characteristics of circZNF609 in human non-malignant diseases.
Focal segmental glomerulosclerosis
Focal segmental glomerulosclerosis (FSGS) is a type of kidney disease, which is characterized by scar tissue that forms in some of the glomeruli in the kidney. FSGS ranks as the first most common cause of adult nephrotic syndrome in the USA and fourth in China [Citation119,Citation120]. Some studies have revealed that the pathogenesis of FSGS is related to protein loss resulting from podocyte damage and depletion and subsequent development of focal sclerosing lesions [Citation121]. Hence, it is pivotal to explore the pathogenesis of FSGS to improve the cure rate of FSGS and reduce the incidence of serious complications.
The interference of circZNF609 was found to play a significant role in the mechanism of FSGS [Citation114]. The expression of circZNF609 was increased while miR-615-5p was downregulated in FSGS biopsies. It was found that renal circZNF609 was positively correlated with proteinuria, serum cholesterol and BUN, and negatively correlated with serum albumin. In addition, the outcome of the study demonstrated that expression of podocyte-specific protein WT1 was decreased and fibrotic protein COL1 was increased in FSGS biopsies [Citation114]. Consequently, circZNF609 may play a crucial role in the pathogenesis of FSGS by reducing podocyte biomarker WT1 and increasing the production of fibrosis protein COL1 via sponging miR-615-5p, which suggests circZNF609 may be a novel therapeutic target for FSGS.
Stress ulcer
Stress ulcer is a common concern among medical professionals. It can increase complications and mortality, prolong hospital stays and significantly reduce patients’ quality of life [Citation122]. It is suggested that the prevention of stress ulcers and further study of the pathogenesis of stress ulcers are prominent objectives.
Ge et al. [Citation115] found that the expression level of circZNF609 was significantly elevated by H2O2 treatment as compared to control. However, the silencing of circZNF609 alleviated the oxidative stress damage in HaCaT cells initiated by H2O2. What is more, circZNF609 silence remarkably promoted cell viability, repressed p53 and p16 expression, reduced apoptosis and inhibited ROS (reactive oxygen species) generation. The research indicated that circZNF609 silencing may function via regulating miR-145 expression and thereby mediating JNK and p38MAPK pathways, which provides us with a new insight and treatment thought.
Corneal neovascularization
The cornea is a transparent and vascular structure on the outer layer of the eye wall, which plays a critical role in protecting the contents of the eye and maintaining good vision [Citation123]. However, when suffering from many pathological injuries such as infection and inflammation, it will promote corneal neovascularization. Corneal neovascularization is one of the causes of global vision loss, which can lead to corneal opacification, and often leads to chronic inflammatory cycles [Citation124]. Therefore, understanding the molecular mechanisms of corneal angiogenesis and vascular inhibition is critical.
CircZNF609 was recently found to participate in corneal neovascularization [Citation116]. The expression of circZNF609 was significantly increased and miR-184 was dramatically decreased compared to normal controls. Forced expression of miR-184 decreased p-AKT, VEGF, and β-catenin expression levels, leading to suppression of HCEKs cell proliferation and migration, as well as HDMECs tube formation, while this inhibition effect could be rescued by circZNF609. Thus, the anti-neovascularization effect of circZNF609 was through targeting miR-184 mediated AKT/β-catenin/VEGF signaling [Citation116]. Hence, circZNF609 or miR-184 may be a promising novel target for the treatment of pathological corneal neovascularization. Additionally, the study of the mechanism of circZNF609 on corneal neovascularization is limited to rat models and needs further study.
Neuropathic pain
Neuropathic pain is caused by lesions or diseases of the somatosensory system and has a significant impact on quality of life [Citation125]. A variety of risk factors such as diabetes, trauma, cancer and so on can cause neuropathic pain [Citation126]. Owing to the low efficiency of clinical intervention on neuropathic pain, it has become a serious problem for patients and the world health system [Citation127]. Consequently, it is imperative to find a novel therapeutic target for neuropathic pain.
Li et al. [Citation38] found that circZNF609 might participate in neuropathic pain progression in CCI rat models. It was found that the expression of circZNF609 was upregulated in CCI rats. Upregulation of miR-22-3p attenuated neuropathic pain in CCI rats and decreased the TNF-α, IL-1 and IL-6 levels in CCI rats, while overexpression of ENO1 could reverse this inhibitory effect. All in all, circZNF609 mediated the expression of inflammatory factors by regulating the miR-22-3p/ENO1 axis to aggravate neuropathic pain progression in CCI rat models [Citation38]. But whether circZNF609 plays the same role in humans needs further study. Overall, the study provides us with a novel direction for exploring potential neuropathic pain therapy.
Vascular endothelial dysfunction
Vascular endothelial dysfunction is a marker of many malignant, inflammatory and ischemic diseases, which also contribute to disease progression [Citation128]. Therefore, finding targets for the treatment of vascular endothelial dysfunction is of great therapeutic significance for the prevention and treatment of vascular complications [Citation129].
Researchers Liu et al. [Citation39] found that circZNF609 was remarkably upregulated in diabetic retinas. Silencing circZNF609 could decline retinal vessel loss and inhibit pathological angiogenesis in vivo. What is more, knockdown of circZNF609 significantly promote the proliferation, migration and tube formation of HRVECs under basal conditions and protect endothelial cells against hypoxic and oxidative stress in vitro. CircZNF609 mainly regulated endothelial cell function via the circZNF609/miR-615-5p/MEF2A axis, which might provide a novel therapeutic target for treating vascular diseases.
Coronary artery disease
Coronary artery disease (CAD) is a condition in which part or all of the coronary arteries that supply blood to the heart muscle are blocked, which is the leading global cause of mortality [Citation130]. Previous studies have found that inflammation plays a crucial role in the process of atherosclerosis [Citation131,Citation132], thus it is necessary to explore potential inflammation-related biomarkers to predict and monitor CAD.
CircZNF609 might be involved in the pathological process in CAD [Citation117]. The finding discovered that the expression of circZNF609 was downregulated significantly in PBLs of patients compared to controls and was negatively associated with CRP levels. Moreover, bioinformatics analysis results showed that circZNF609 had a moderate predictive value in distinguishing CAD patients. They found that overexpression of circZNF609 could attenuate inflammation and circZNF609 might bind miR-145-5p, miR-615-5p, miR-138-5p, miR-150-5p to exert its function. CircZNF609 might play a role by reducing the intensity of the inflammatory response to function as an independent protective factor and may be used as a new biomarker for the diagnosis of CAD.
Glaucoma
Glaucoma is a progressive retinal neurodegenerative disease characterized by a gradual decrease in retinal ganglion cells (RGCs) [Citation133]. RGCs are the sole output neurons that transmit visual information from the retina to the brain. Different damages and pathological states lead to degeneration of RGC somas and axons, leading to irreversible vision loss [Citation134]. Therefore, an in-depth understanding of the exact mechanism of retinal neurodegeneration in glaucoma is urgently needed.
There was a novel clue that circZNF609 might act an important role through regulating retinal neurodegeneration in glaucoma [Citation118]. Wang et al. [Citation118] found that circZNF609 expression was significantly upregulated during retinal neurodegeneration. CircZNF609 silencing led to decreased cell viability of Müller cells in glaucoma. CircZNF609 regulated retinal neurodegeneration via a circZNF609/miR-615/METRN regulatory network, which provided an encouraging potential target for treating retinal neurodegeneration [Citation118].
Future outlook
For a long time, scientists have explored new cancer treatment strategies, and the discovery of circRNAs has offered new hope. This paper summarizes the current research progress about circRNA circZNF609, which has been newly discovered so far, according to the classification of eight human systems, which may deepen our understanding of how it regulates tumor development.
Researchers have shown that circZNF609 could be used as a biomarker to predict tumor progression and disease prognosis. The expression of circZNF609 is upregulated in various malignant tumors, including lung cancer [Citation50,Citation51, Citation61], CRC [Citation68], NPC [Citation46, Citation63–65], glioma [Citation43,Citation44], renal carcinoma [Citation71], CC [Citation74], HCC [Citation47, Citation67], RMS [Citation45] and melanoma [Citation61], to name a few. In addition, the abnormal expression of circZNF609 in tumor tissues closely correlates with lymph node metastasis, advanced TNM stage and poor prognosis. Meanwhile, circZNF609 has been found to significantly enhance cell viability and proliferation, promote cell invasion and migration and inhibit cell apoptosis in most experimental settings. In terms of function, circZNF609 mainly acts as a miRNA molecular sponge, competitively binding miRNAs to modulate multiple cancer-related proteins, such as ETV1, FOXM1 and GNB2. It is worth noting that, in CRC, the changes in circZNF609 expression appear to be contradictory, according to different reports. Wu et al. [Citation68] reported that circZNF609 expression was upregulated in CRC, while Zhang et al. [Citation69] observed that it was downregulated. We believe that this contradiction might result from an uneven selection of samples by the authors. Wu et al. [Citation68] reported that CRC tissue samples were mainly obtained from patients with CRC who had undergone surgical resection. However, the samples used in the study by Zhang et al. [Citation69] were collected from the peripheral blood before surgery. Thus, the expression and chemical stability of circZNF609 in biological samples such as plasma, serum and serum-derived exosomes have not been demonstrated in CRC and require further investigation.
At the same time, a recent study found that circZNF609 can also interact with CKAP5 mRNA to down-regulate the level of CKAP5 protein, which in turn leads to dysregulation of microtubule dynamics, affects the mitosis process and damages DNA, so circZNF609 plays a certain role in anti-tumor drugs of MT-targeted drugs. Regardless of tumor histological origin or corresponding miRNA signature, the regulation of the ubiquitous CKAP5 protein in cells by circZNF609 can play a role in circZNF609-overexpressed tumor cells. Therefore, the mechanism of circZNF609-mRNA interaction may need to be further studied to bring more hope for clinical treatment [Citation52,Citation53].
In addition, the discovery of circZNF609 also brings good news for the treatment of non-cancer diseases. In HSCR, circZNF609 regulates AKT3 expression by acting as a sponge for miR-150-5p, thereby participating in the development of HSCR [Citation113]. CircZNF609 is also involved in the abnormal vascular expression, and depletion of circZNF609 promotes the survival, and proliferation of vascular endothelial cells, and protects endothelial cells from hypoxia and oxidative stress in vitro [Citation39]. In addition, circZNF609 can also regulate glaucoma retinal neurodegeneration by affecting the activity and proliferation ability of Müller cells and RGC cells [Citation118]. What is more, overexpression of circZNF609 could act as a protective factor by attenuating inflammation in CAD [Citation117].
Conclusions
Although great strides have been made in understanding the function of circZNF609 in various human diseases, since the current sample size and experimental data are not sufficient to provide strong supporting evidence, further experiments are urgently needed to explore the functional roles and molecular mechanisms of circZNF609 in human diseases. To date, circZNF609 has been found to act as a molecular sponge in human diseases, interact with mRNAs, encode proteins and perform as a therapeutic target. Its role in human diseases is yet to be studied further. For example, the relation between circZNF609 and tumor microenvironment, cancer stem cells, cancer immune escape, will be the directions of future research on circZNF609. In addition, circZNF609 contains a 753-nt open reading frame that can be translated into protein. The functional role of the translation protein of circZNF609, whether it can be used as a protein chaperone, and other more non-ceRNA mechanisms are research topics worthy of further study. Overall, we conclude that circZNF609 is a potential biomarker, which can predict human disease development and provide clinical guidance for human disease treatment.
Statements and declarations
Consent for publication
All authors consent to publish the article in this journal.
Author contributions
Minhua Wu and Xiaoxia Ye designed the study. Jieyi Luo and Shengchun Wang wrote the first draft of the manuscript. Lu Zhang and Shanshan Wu drafted the picture. Weirang Zheng and Xueshan Huang revised the manuscript. All authors approved the final manuscript.
Ethics approval
Not applicable
Consent to participate
Not applicable
Disclosure statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Data availability
All data generated or analyzed during this study are included in this article.
Correction statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Chen L-L, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388.
- Wu H, Yang L, Chen L-L. The diversity of long noncoding RNAs and their generation. Trends Genet. 2017;33(8):540–552.
- Zhai X, Zhang Y, Xin S, et al. Insights into the involvement of circular RNAs in autoimmune diseases. Front Immunol. 2021;12:622316.
- Yang X, Ye T, Liu H, et al. Expression profiles, biological functions and clinical significance of circRNAs in bladder cancer. Mol Cancer. 2021;20(1):4.
- Cocquerelle C, Mascrez B, Hétuin D, et al. Mis-splicing yields circular RNA molecules. Faseb J. 1993;7(1):155–160.
- Nigro JM, Cho KR, Fearon ER, et al. Scrambled exons. Cell. 1991;64(3):607–613.
- Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264.
- Liu X, Hu Z, Zhou J, et al. Interior circular RNA. RNA Biol. 2020;17(1):87–97.
- Rodríguez-Trelles F, Tarrío R, Ayala F. Origins and evolution of spliceosomal introns. Annu Rev Genet. 2006;40:47–76.
- Zhao X, Cai Y, Xu J. Circular RNAs: biogenesis, mechanism, and function in human cancers. IJMS. 2019;20(16):3926.
- Zhou W-Y, Cai Z-R, Liu J, et al. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19(1):172.
- Wen G, Zhou T, Gu W. The potential of using blood circular RNA as liquid biopsy biomarker for human diseases. Protein Cell. 2021;12(12):911–946.
- Shi X, Wang B, Feng X, et al. circRNAs and exosomes: a mysterious frontier for human cancer. Mol Ther Nucleic Acids. 2020;19:384–392.
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA (New York, N.Y.). 2013;19(2):141–157.
- Huang C, Liang D, Tatomer DC, et al. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018;32(9-10):639–644.
- Chen L-L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21(8):475–490.
- Li J-q, Yang J, Zhou P[, et al. The biological functions and regulations of competing endogenous RNA. ]Yi Chuan. 2015;37(8):756–764.
- Huang A, Zheng H, Wu Z, et al. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics. 2020;10(8):3503–3517.
- Chen C, Yang Y, Wang Z. Study of circular RNA translation using reporter systems in living cells. Methods (San Diego, Calif.). 2021;196:113–120.
- van Zonneveld AJ, Kölling M, Bijkerk R, et al. Circular RNAs in kidney disease and cancer. Nat Rev Nephrol. 2021;17(12):814–826.
- Abe N, Matsumoto K, Nishihara M, et al. Rolling circle translation of circular RNA in living human cells. Sci Rep. 2015;5:16435.
- Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell. 2017;66(1):9–21.e7. e27.
- Begum S, Yiu A, Stebbing J, et al. Novel tumour suppressive protein encoded by circular RNA, circ-SHPRH, in glioblastomas. Oncogene. 2018;37(30):4055–4057.
- Zhang Y, Jiang J, Zhang J, et al. CircDIDO1 inhibits gastric cancer progression by encoding a novel DIDO1-529aa protein and regulating PRDX2 protein stability. Mol Cancer. 2021;20(1):101.
- Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66(1):22–37.e9. e29.
- Xiao M-S, Wilusz JE. An improved method for circular RNA purification using RNase R that efficiently removes linear RNAs containing G-quadruplexes or structured 3’ ends. Nucleic Acids Res. 2019;47(16):8755–8769.
- Knupp D, Cooper DA, Saito Y, et al. NOVA2 regulates neural circRNA biogenesis. Nucleic Acids Res. 2021;49(12):6849–6862.
- Zhang J, Zhao F. Reconstruction of circular RNAs using illumina and nanopore RNA-seq datasets. Methods. 2021;196:17–22.
- Zhang J, Hou L, Zuo Z, et al. Comprehensive profiling of circular RNAs with nanopore sequencing and CIRI-long. Nat Biotechnol. 2021;39(7):836–845.
- Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94.
- Bai Y, Zhang Y, Han B, et al. Circular RNA DLGAP4 ameliorates ischemic stroke outcomes by targeting miR-143 to regulate endothelial-mesenchymal transition associated with blood-brain barrier integrity. J Neurosci. 2018;38(1):32–50.
- Wu P, Mo Y, Peng M, et al. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol Cancer. 2020;19(1):22.
- Geng Y, Jiang J, Wu C. Function and clinical significance of circRNAs in solid tumors. J Hematol Oncol. 2018;11(1):98.
- Guarnerio J, Bezzi M, Jeong JC, et al. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;166(4):1055–1056.
- Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. 2021;21(1):22–36.
- Lux S, Bullinger L. Circular RNAs in cancer. Adv Exp Med Biol. 2018;1087:215–230.
- Rybak-Wolf A, Stottmeister C, Glazar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870–885.
- Li L, Luo Y, Zhang Y, et al. CircZNF609 aggravates neuropathic pain via miR-22-3p/ENO1 axis in CCI rat models. Gene. 2020;763:145069.
- Liu C, Yao MD, Li CP, et al. Silencing of circular RNA-ZNF609 ameliorates vascular endothelial dysfunction. Theranostics. 2017;7(11):2863–2877.
- Wang S, Xue X, Wang R, et al. CircZNF609 promotes breast cancer cell growth, migration, and invasion by elevating p70S6K1 via sponging miR-145-5p. CMAR. 2018;ume 10:3881–3890.
- Liu Z, Pan HM, Xin L, et al. Circ-ZNF609 promotes carcinogenesis of gastric cancer cells by inhibiting miRNA-145-5p expression. Eur Rev Med Pharmacol Sci. 2019;23(21):9411–9417.
- Wu W, Wei N, Shao G, et al. circZNF609 promotes the proliferation and migration of gastric cancer by sponging miR-483-3p and regulating CDK6. Onco Targets Ther. 2019;12:8197–8205.
- Du S, Li H, Lu F, et al. Circular RNA ZNF609 promotes the malignant progression of glioma by regulating miR-1224-3p/PLK1 signaling. J Cancer. 2021;12(11):3354–3366.
- Tong H, Zhao K, Wang J, et al. CircZNF609/miR-134-5p/BTG-2 axis regulates proliferation and migration of glioma cell. J Pharm Pharmacol. 2020;72(1):68–75.
- Rossi F, Legnini I, Megiorni F, et al. Circ-ZNF609 regulates G1-S progression in rhabdomyosarcoma. Oncogene. 2019;38(20):3843–3854.
- Wang J, Lin Y, Jiang DH, et al. CircRNA ZNF609 promotes angiogenesis in nasopharyngeal carcinoma by regulating miR-145/STMN1 axis. Kaohsiung J Med Sci. 2021;37(8):686–698.
- He Y, Huang H, Jin L, et al. CircZNF609 enhances hepatocellular carcinoma cell proliferation, metastasis, and stemness by activating the hedgehog pathway through the regulation of miR-15a-5p/15b-5p and GLI2 expressions. Cell Death Dis. 2020;11(5):358.
- Yang H, Li X, Meng Q, et al. CircPTK2 (hsa_circ_0005273) as a novel therapeutic target for metastatic colorectal cancer. Mol Cancer. 2020;19(1):13.
- Li D, Li Z, Yang Y, et al. Circular RNAs as biomarkers and therapeutic targets in environmental chemical exposure-related diseases. Environ Res. 2020;180:108825.
- Liu S, Yang N, Jiang X, et al. FUS-induced circular RNA ZNF609 promotes tumorigenesis and progression via sponging miR-142-3p in lung cancer. J Cell Physiol. 2021;236(1):79–92.
- Wang F, Li X, Jia X, et al. CircRNA ZNF609 knockdown represses the development of Non-Small cell lung cancer via miR-623/FOXM1 axis. Cancer Manag Res. 2021;13:1029–1039.
- Beltran M, Rossi F, Bozzoni I. CircZNF609 as a prototype to elucidate the biological function of circRNA-mRNA interactions. Mol Cell Oncol. 2022;9(1):2055939.
- Rossi F, Beltran M, Damizia M, et al. Circular RNA ZNF609/CKAP5 mRNA interaction regulates microtubule dynamics and tumorigenicity. Mol Cell. 2022;82(1):75–89.e9. e9.
- Thawani A, Kadzik RS, Petry S. XMAP215 is a microtubule nucleation factor that functions synergistically with the γ-tubulin ring complex. Nat Cell Biol. 2018;20(5):575–585.
- Miller MP, Asbury CL, Biggins S. A TOG protein confers tension sensitivity to kinetochore-microtubule attachments. Cell. 2016;165(6):1428–1439.
- Di Timoteo G, Dattilo D, Centrón-Broco A, et al. Modulation of circRNA metabolism by mA modification. Cell Rep. 2020;31(6):107641.
- Ho-Xuan H, Glažar P, Latini C, et al. Comprehensive analysis of translation from overexpressed circular RNAs reveals pervasive translation from linear transcripts. Nucleic Acids Res. 2020;48(18):10368–10382.
- Ostrom QT, Bauchet L, Davis FG, et al. The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol. 2014;16(7):896–913.
- Ghouzlani A, Kandoussi S, Tall M, et al. Immune checkpoint inhibitors in human glioma microenvironment. Front Immunol. 2021;12:679425–679425.
- Zhao Z, Li G, Han Y, et al. Circular RNA ZNF609 enhances proliferation and glycolysis during glioma progression by miR-378b/SLC2A1 axis. Aging (Albany NY). 2021;13(17):21122–21133.
- Zuo Y, Shen W, Wang C, et al. Circular RNA Circ-ZNF609 promotes lung adenocarcinoma proliferation by modulating miR-1224-3p/ETV1 signaling. Cancer Manag Res. 2020;12:2471–2479.
- Yin X, Wang J, Shan C, et al. Circular RNA ZNF609 promotes laryngeal squamous cell carcinoma progression by upregulating epidermal growth factor receptor via sponging microRNA-134-5p. Bioengineered. 2022;13(3):6929–6941.
- Li M, Li Y, Yu M. CircRNA ZNF609 knockdown suppresses cell growth via modulating miR-188/ELF2 axis in nasopharyngeal carcinoma. Onco Targets Ther. 2020;13:2399–2409.
- Zhu L, Liu Y, Yang Y, et al. CircRNA ZNF609 promotes growth and metastasis of nasopharyngeal carcinoma by competing with microRNA-150-5p. Eur Rev Med Pharmacol Sci. 2019;23(7):2817–2826.
- Liu Z, Liu F, Wang F, et al. CircZNF609 promotes cell proliferation, migration, invasion, and glycolysis in nasopharyngeal carcinoma through regulating HRAS via miR-338-3p. Mol Cell Biochem. 2021;476(1):175–186.
- Yang CS, Lou Y, Ke QP[, et al. Mechanism of circZNF609 targeting miR-153 to regulate the proliferation and apoptosis of diffuse large B-cell lymphoma. ]. Zhonghua Zhong Liu Za Zhi. 2022;44(3):238–245.
- Liao X, Zhan W, Tian B, et al. Circular RNA ZNF609 promoted hepatocellular carcinoma progression by upregulating PAP2C expression via sponging miR-342-3p. Onco Targets Ther. 2020;13:7773–7783.
- Wu L, Xia J, Yang J, et al. Circ-ZNF609 promotes migration of colorectal cancer by inhibiting Gli1 expression via microRNA-150. J BUON: Off J Balkan Union Oncol. 2018;23(5):1343–1349.
- Zhang X, Zhao Y, Kong P, et al. Expression of circZNF609 is down-regulated in colorectal cancer tissue and promotes apoptosis in colorectal cancer cells by upregulating p53. Med Sci Monit. 2019;25:5977–5985.
- Guan C, Liu L, Zhao Y, et al. YY1 and eIF4A3 are mediators of the cell proliferation, migration and invasion in cholangiocarcinoma promoted by circ-ZNF609 by targeting miR-432-5p to regulate LRRC1. Aging (Albany NY). 2021;13(23):25195–25212. )
- Xiong Y, Zhang J, Song C. CircRNA ZNF609 functions as a competitive endogenous RNA to regulate FOXP4 expression by sponging miR-138-5p in renal carcinoma. J Cell Physiol. 2019;234(7):10646–10654.
- Jin C, Zhao W, Zhang Z, et al. Silencing circular RNA circZNF609 restrains growth, migration and invasion by up-regulating microRNA-186-5p in prostate cancer. Artif Cells Nanomed Biotechnol. 2019;47(1):3350–3358.
- Du S, Zhang P, Ren W, et al. Circ-ZNF609 accelerates the radioresistance of prostate cancer cells by promoting the glycolytic metabolism through miR-501-3p/HK2 axis. Cancer Manag Res. 2020;12:7487–7499.
- Gu Q, Hou W, Shi L, et al. Circular RNA ZNF609 functions as a competing endogenous RNA in regulating E2F transcription factor 6 through competitively binding to microRNA-197-3p to promote the progression of cervical cancer progression. Bioengineered. 2021;12(1):927–936.
- Shi P, Liu Y, Yang D, et al. CircRNA ZNF609 promotes the growth and metastasis of thyroid cancer and by downregulating miR-514a-5p. Bioengineered. 2022;13(2):4372–4384.
- Liu Q, Cui W, Yang C, et al. Circular RNA ZNF609 drives tumor progression by regulating the miR-138-5p/SIRT7 axis in melanoma. Aging (Albany NY). 2021;13(15):19822–19834.
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30.
- Campion NJ, Ally M, Jank BJ, et al. The molecular march of primary and recurrent nasopharyngeal carcinoma. Oncogene. 2021;40(10):1757–1774.
- Guo L-L, Wang H-Y, Zheng L-S, et al. Metastasis of nasopharyngeal carcinoma: what we know and do not know. Vis Cancer Med. 2021;2:4.
- Li S, Young KH, Medeiros LJ. Diffuse large B-cell lymphoma. Pathology. 2018;50(1):74–87.
- Craig AJ, von Felden J, Garcia-Lezana T, et al. Tumour evolution in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2020;17(3):139–152.
- Llovet JM, Zucman-Rossi J, Pikarsky E, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2(1):16018.
- Rawla P, Barsouk A. Epidemiology of gastric cancer: global trends, risk factors and prevention. Prz Gastroenterol. 2019;14(1):26–38.
- Janney A, Powrie F, Mann EH. Host-microbiota maladaptation in colorectal cancer. Nature. 2020;585(7826):509–517.
- Aran V, Victorino AP, Thuler LC, et al. Colorectal cancer: epidemiology, disease mechanisms and interventions to reduce onset and mortality. Clin Colorectal Cancer. 2016;15(3):195–203.
- Ho-Xuan H, Lehmann G, Glazar P, et al. Gene expression signatures of a preclinical mouse model during colorectal cancer progression under low-dose metronomic chemotherapy. Cancers (Basel). 2020;13(1):49.
- Saha SK, Zhu AX, Fuchs CS, et al. Forty-Year trends in cholangiocarcinoma incidence in the U.S. Oncologist. 2016;21(5):594–599.
- Shelar S, Shim E-H, Brinkley GJ, et al. Biochemical and epigenetic insights into L-2-hydroxyglutarate, a potential therapeutic target in renal cancer. Clin Cancer Res. 2018;24(24):6433–6446.
- Chen P, Pan X, Zhao L, et al. MicroRNA-191-5p exerts a tumor suppressive role in renal cell carcinoma. Exp Ther Med. 2018;15(2):1686–1693.
- Capitanio U, Montorsi F. Renal cancer. Lancet. 2016;387(10021):894–906.
- Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clin. 2021;71(3):209–249.
- Chen R-J, Hung C-M, Chen Y-L, et al. Monascuspiloin induces apoptosis and autophagic cell death in human prostate cancer cells via the Akt and AMPK signaling pathways. J Agric Food Chem. 2012;60(29):7185–7193.
- Sheng X, Li W-B, Wang D-L, et al. Yap is closely correlated with castration-resistant prostate cancer, and downregulation of Yap reduces proliferation and induces apoptosis of PC-3 cells. Mol Med Rep. 2015;12(4):4867–4876.
- Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464.
- Zhang Y, Xu Z, Ding J, et al. HZ08 suppresses RelB-activated MnSOD expression and enhances radiosensitivity of prostate cancer cells. J Exp Clin Cancer Res. 2018;37(1):174.
- Fotouhi Ghiam A, Taeb S, Huang X, et al. Long non-coding RNA urothelial carcinoma associated 1 (UCA1) mediates radiation response in prostate cancer. Oncotarget. 2017;8(3):4668–4689.
- Angahar LT. An overview of breast cancer epidemiology, risk factors, pathophysiology, and cancer risks reduction. MOJBM. 2017;1(4):92–96.
- Wen Y, Zhang S, Yang J, et al. Identification of driver genes regulating immune cell infiltration in cervical cancer by multiple omics integration. Biomed Pharmacother. 2019;120:109546.
- Goodman A. HPV testing as a screen for cervical cancer. BMJ. 2015;350:h2372.
- Kitahara CM, Sosa JA. The changing incidence of thyroid cancer. Nat Rev Endocrinol. 2016;12(11):646–653.
- Kim J, Gosnell JE, Roman SA. Geographic influences in the global rise of thyroid cancer. Nat Rev Endocrinol. 2020;16(1):17–29.
- Xing M. Molecular pathogenesis and mechanisms of thyroid cancer. Nat Rev Cancer. 2013;13(3):184–199.
- Almacellas-Rabaiget O, Monaco P, Huertas-Martinez J, et al. LOXL2 promotes oncogenic progression in alveolar rhabdomyosarcoma independently of its catalytic activity. Cancer Lett. 2020;474:1–14.
- Skapek SX, Ferrari A, Gupta AA, et al. Rhabdomyosarcoma. Nat Rev Dis Primers. 2019;5(1):1–1.
- Sun X, Guo W, Shen JK, et al. Rhabdomyosarcoma: advances in molecular and cellular biology. Sarcoma. 2015;2015:232010.
- Egas-Bejar D, Huh WW. Rhabdomyosarcoma in adolescent and young adult patients: current perspectives. AHMT. 2014;5:115–125.
- Giugliano F, Crimini E, Tarantino P, et al. First line treatment of BRAF mutated advanced melanoma: does one size fit all? Cancer Treat Rev. 2021;99:102253.
- Marzagalli M, Ebelt ND, Manuel ER. Unraveling the crosstalk between melanoma and immune cells in the tumor microenvironment. Semin Cancer Biol. 2019;59:236–250.
- Paluncic J, Kovacevic Z, Jansson PJ, et al. Roads to melanoma: key pathways and emerging players in melanoma progression and oncogenic signaling. Biochim Biophys Acta. 2016;1863(4):770–784.
- Olive PL, Banáth JP. The comet assay: a method to measure DNA damage in individual cells. Nat Protoc. 2006;1(1):23–29.
- Arshad A, Powell C, Tighe MP. Hirschsprung’s disease. BMJ. 2012;345:e5521.
- Peng L, Chen G, Zhu Z, et al. Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in hirschsprung’s disease. Oncotarget. 2017;8(1):808–818.
- Cui X, Fu J, Luan J, et al. CircZNF609 is involved in the pathogenesis of focal segmental glomerulosclerosis by sponging miR-615-5p. Biochem Biophys Res Commun. 2020;531(3):341–349.
- Ge R, Gao G. Anti-antioxidant impacts of circZNF609 silence in HaCaT cells through regulating miR-145. Artif Cells Nanomed Biotechnol. 2020;48(1):384–392.
- Wu P, Zhang D, Geng Y, et al. Circular RNA-ZNF609 regulates corneal neovascularization by acting as a sponge of miR-184. Exp Eye Res. 2020;192:107937.
- Liang B, Li M, Deng Q, et al. CircRNA ZNF609 in peripheral blood leukocytes acts as a protective factor and a potential biomarker for coronary artery disease. Ann Transl Med. 2020;8(12):741–741.
- Wang JJ, Liu C, Shan K, et al. Circular RNA-ZNF609 regulates retinal neurodegeneration by acting as miR-615 sponge. Theranostics. 2018;8(12):3408–3415.
- Rosenberg AZ, Kopp JB. Focal segmental glomerulosclerosis. Clin J Am Soc Nephrol. 2017;12(3):502–517.
- Xu X, Wang G, Chen N, et al. Long-Term exposure to air pollution and increased risk of membranous nephropathy in China. JASN. 2016;27(12):3739–3746.
- Wiggins RC. The spectrum of podocytopathies: a unifying view of glomerular diseases. Kidney Int. 2007;71(12):1205–1214.
- Agrawal K, Chauhan N. Pressure ulcers: back to the basics. Indian J Plast Surg. 2012;45(2):244–254.
- Meek KM, Knupp C. Corneal structure and transparency. Prog Retin Eye Res. 2015;49:1–16.
- Nicholas MP, Mysore N. Corneal neovascularization. Exp Eye Res. 2021;202:108363.
- Finnerup NB, Kuner R, Jensen TS. Neuropathic pain: from mechanisms to treatment. Physiol Rev. 2021;101(1):259–301.
- Gilron I, Baron R, Jensen T. Neuropathic pain: principles of diagnosis and treatment. Mayo Clin Proc. 2015;90(4):532–545.
- van Hecke O, Austin SK, Khan RA, et al. Neuropathic pain in the general population: a systematic review of epidemiological studies. Pain. 2014;155(4):654–662.
- Puro DG, Kohmoto R, Fujita Y, et al. Bioelectric impact of pathological angiogenesis on vascular function. Proc Natl Acad Sci U S A. 2016;113(35):9934–9939.
- Qian Y, Li Y, Li R, et al. circ-ZNF609: a potent circRNA in human cancers. J Cell Mol Med. 2021;25(22):10349–10361.
- Khera AV, Kathiresan S. Genetics of coronary artery disease: discovery, biology and clinical translation. Nat Rev Genet. 2017;18(6):331–344.
- Tardif J-C, Kouz S, Waters DD, et al. Efficacy and safety of low-dose colchicine after myocardial infarction. N Engl J Med. 2019;381(26):2497–2505.
- Ridker PM, Everett BM, Thuren T, CANTOS Trial Group, et al. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med. 2017;377(12):1119–1131.
- Almasieh M, Wilson AM, Morquette B, et al. The molecular basis of retinal ganglion cell death in glaucoma. Prog Retin Eye Res. 2012;31(2):152–181.
- Guo X, Zhou J, Starr C, et al. Preservation of vision after CaMKII-mediated protection of retinal ganglion cells. Cell. 2021;184(16):4299–4314.e12.
