Abstract
Bacillus coagulans LYBC06 strain with lactate-producing capacity was isolated from mucosal tissue of the digestive tract of a fattening pig. The resequencing of the LYBC06 strain revealed that the overall genome was less variable, with a genome-wide heterozygous ratio of 0.042 %, total SNP locus variants 0.4273% of the entire genome, total InDel mutations 0.0182%, total increased and decreased CNV copies 0.0031% and total SV annotated variants 0.0083% of the entire genome. The mutated genes were mainly involved in the growth of the strain, regulation of energy metabolism, and production of proteases and lactate. The whole genome of LYBC06 strain was sequenced with the length of 3385106 bp, GC content of 46.86% and 4092 coding genes (85.96% of coding). Functional annotation of the proteins encoding genes showed that the growth process of LYBC06 strain involved several metabolic pathway transformation processes of various substances, and many encoding genes were involved in the resistance to macrolides, polypeptides and tetracyclines. The basic characteristics of the genome of LYBC06 strain were relatively similar to those of other Bacillus coagulans, and the collinearity between several Bacillus coagulans strains was relatively good. Bacterial growth, gene expression of lactate dehydrogenase and lactate production in LYBC06 strain were largely synchronized, and the maximum amount of lactate could be produced up to 92 g/L at 16 h culture during the logarithmic growth phase, in which the LdhL1 gene played an important role in the production of lactate in LYBC06 strain.
Introduction
The restricted use of antibiotics in livestock feed has become a consensus of the international community. Since July 1, 2020, a comprehensive prohibition of antibiotics in feeds has been implemented in China. The protection of livestock health has become a big challenge in our country, and microecological agents are the perfect substitutes [Citation1]. Microecological preparation can not only regulate the balance of flora and reduce the number of Escherichia coli in piglet feces, but can also promote the absorption and metabolism of nutrients and improve the utilization of feeds [Citation2]. Bacillus coagulans is one of the 18 safe microecological strains allowed for addition in feed as per the announcement of the Ministry of Agriculture of China in 2012. The bacterium belongs to the gram-positive bacilli with spore-producing capacity. It is a thermophilic homotypic lactic acid-fermenting microorganism, which can produce a large amount of lactic acid and belongs to the so-called ‘spore-forming lactic acid bacteria’ in the intestines [Citation3]. Studies have shown that direct utilization of B. coagulans fermentation without sterilization under 50 °C culture conditions can produce l-lactic acid with more than 99% optical purity and high potential for commercial applications [Citation4]. B. coagulans functions as a general lactic acid bacterium to maintain the balance of intestinal microecology in animals, and it can improve animal body immunity and animal reproduction performance [Citation5]. Meanwhile, B. coagulans, as a member of genus Bacillus, has attracted much attention in the bio-feed industry in the recent years due to its excellent characteristics, such as the capability to generate spores, resistance to strong stress, high temperature and storage, and ability to facilitate feed processing [Citation6]. B. coagulans is a facultative anaerobe, with a relatively lower requirement for growth environment. B. coagulans can tolerate strong acid and high bile salt environment of the simulated gastrointestinal tract, thereby having the potential to serve as a probiotic. On the other hand, its capability to generate spores directly enhances its stress resistance offering tolerance to high temperature during feed processing, convenient storage, and so on. Compared with probiotics such as Bifidobacterium sp. and Lactobacillus acidophilus, B.coagulans has more advantages such as fast growth and strong stress resistance [Citation7,Citation8].
Currently, although the safety, prebiotic potential and importance of B. coagulans have been recognized, it has not been widely used in the feed industry so far, and its excellent biological characteristics have not been fully exploited. One of the major reasons is that its genome-wide information, especially the information of its lactic acid fermentation-related genes, is not fully understood and analyzed [Citation9]. Therefore, in this study, a B. coagulans strain with strong stress resistance and bacteriostatic activity was isolated from the gastrointestinal tracts of healthy pigs. The strain was screened to exert its ability to adjust the intestinal health of animals during animal culture and replace antibiotics by inhibiting harmful bacteria, which will provide a new hope of obtaining superior species that can be applied in feed production practice. This study also applied the combination of the second-generation sequencing technology based on the Illumina MiSeq sequencing platform and the third-generation single-molecule sequencing technology based on the PacBioRs II sequencing platform to perform full gene sequencing analysis of the strain, in the hope of providing a comprehensive analysis of the genomic information of B. coagulans and a reference for a deeper understanding of the relationship between transcription of lactate dehydrogenase gene and lactate synthesis in the bacterium. This study also provides a theoretical basis for the applications of B. coagulans in the fields of animal microecological feed additives, food industries and health products.
Materials and methods
Sample source for isolating the Bacillus coagulans strain
The large intestine, small intestine and rectum of digestive tracts from a fattening DLY (Duroc × Landrace × Yorkshire) pig with body weight of 60 kg, raised in a pig farm of Xingyue Ecological Breeding Co., Ltd., Longyan City, China.
Medium
For isolation and seed culture of lactic acid bacteria, we used MRS (Man Rogosa Sharpe Medium), which consisted of 10 g/L peptone, 5 g/L beef extract powder, 20 g/L glucose, 1 mL/L Tween 80, 2 g/L KH2PO4·7H2O, 5 g/L CH3COONa·3H2O, 2 g/L citric acid triamine, 0.2 g/L MgSO4·7H2O, 0.05 g/L MnSO4·7H2O, 20 g/L agar (used with solid medium) and pH 6.2.
Lactic acid bacteria fermentation medium consisted of 90 g/L glucose, 13.33 g/L yeast extract, 0.0133 g/L peptone, 0.0133 g/L MnSO4, 0.0133 g/L MgSO4, 0.0133 g/L FeSO4, 0.0133 g/L NaCl, 0.67 g/L sodium acetate, 20 g/L calcium carbonate and pH 7 0.
Reagents and instruments
We used Lactate dehydrogenase Kit, purchased from Nanjing Jiancheng Bioengineering Co., Ltd., China, for the determination of lactic acid content in samples.
Sequencer (model 3730XL) and Thermal Cycler (model 2720) were all purchased from Applied Biosystems Inc. (USA). Plate centrifuge (model 5810 R) was purchased from Eppendorf Inc. (Germany). Gel imaging device (model JY04S-3C) and Electrophoresis instrument (model JY300C Power Supply) were all purchased from Beijing Junyi Dongfang electrophoresis equipment Co., Ltd., China.
Screening of the strain
On a sterile operation table, the mucosal tissue of digestive tract from the large intestine, small intestine and rectum of a hypertrophic pig was scraped with a scalpel separately, placed in a Petri dish, sheared and minced with scissors, and mixed evenly. One gram of mixed sample was weighted, transferred into a test tube containing 9 mL of normal saline, mixed well and heated in a water bath at 80 °C for 20 min. After heating, 1 mL of mixture was used for gradient dilution to 10−4 and 10−5. Then 0.1 mL of diluted solution was taken and spread into MRS solid medium, placed in a candling cylinder, and incubated at 37 °C for 24–48 h, and the suspected colonies with vigorous growth and good growth potential were isolated for further identification and screening [Citation10].
Analysis for morphology of the selected strain, 16S rDNA and sporulation characteristics
Morphological identification: The observation of plate colonies, Gram stains and spore staining.
16S rDNA sequence analysis: Total bacterial DNA was extracted and purified using 16S universal primers, and 2× Tsingke Master Mix (code No.: TSE003) system was used for polymerase chain reaction (PCR) amplification using dH2O as a negative control. PCR primers were designed as follows: 27 F: 5′-AGAGTTTGATCMTGGCTCAGC-3′; 1492 R: 5′-GGTTACCTTGTTACGACTT-3′ [Citation11].The total PCR reaction volume was 50 µL, including 1.0 µL of DNA template, 25.0 µL of 2× TsingKE Master Mix, 1.0 µL of Primer F (10 µM), 1.0 µL of Primer R (10 µM) and 22.0 µL of ddH2O. The PCR program was: 94 °C for 10 min, 94 °C for 30 s, 55 °C for 30 s, 72 °C for 90 s, with an extension at 72 °C for 10 min after 30 cycles. Approximately 2.0 µL of PCR-amplified products were subjected to 1% agarose gel electrophoresis. The bands were purified by gel recovery and sent to Beijing Allvison Gene Technology Co., Ltd. for sequencing. The 16S rDNA sequences were compared in the NCBI database, the sequence information of relevant strains was downloaded, the phylogenetic evolutionary tree was reconstructed using the software Mega version 6, and the taxonomic status of the strains was analyzed, while the obtained strain was submitted to the China General Microorganism Culture Collection and Management Center for preservation.
Re-sequencing and analysis of the screened strain
The selected strain was re-sequenced by next generation sequencing technology (NGS) to analyze the genetic variation information including single nucleotide polymorphism (SNP), InDels (Insertions or Deletions), structural variations (SV) and copy number variations (CNV) [Citation12].
Whole genome sequencing and functional analysis of the screened strain
The total bacterial genomic DNA of the selected strain was extracted by the CTAB method with reference to Molecular Cloning: A Laboratory Manual (Fourth Edition) [Citation13]. Whole-genome sequencing was completed by the combination of second- and third-generation sequencing technologies based on Illumina MiSeq and PacBioRS II sequencing platforms from Beijing Allvison Gene Technology Co., Ltd. The obtained sequences were corrected and spliced to obtain a complete sequence. A complete genome circle map was constructed and the protein function of the coding gene was annotated. The specific data processing method, genome circle drawing and functional annotation method of the genes and proteins encoded by the studied genome sequence were performed as described by Lin et al. [Citation14].The predicted gene protein coding sequences were input into the GO (Gene Ontology), KEGG (Kyoto Encyclopedia of Genes and Genomes), COG (Clusters of Orthologous Groups for Eukaryotic Complete Genomes), NR (Non-redundant proteins sequence database), Pfam, TCDB (The Transporter Classification Database) and Swiss-Prot (Swiss-Prot protein sequence) databases for alignment (Diamond, Evalue≤1e−5) to analyze the metabolic pathways and metabolic function types of the isolated strain.
Analysis of lactate dehydrogenase-related genes: The basic situation of the lactate dehydrogenase-related genes was summarized by searching the genome-wide information of the assayed strain.
Comparative genomic analysis of screened strains
Other sequenced genome sequence information for B.coagulans was downloaded from the GenBank database for comparative analysis with the genome information of the strain screened in this study, including basic characteristics, consensus and specific genes of each strain. Genomic collinearity analysis was performed using Mauve software [Citation15] to analyze the differences in the distribution of functional genes between the screened strain and other sequenced species. It provides a theoretical basis for studying the evolutionary process of B. coagulans genome.
Analysis for lactate-producing characteristics and gene expression of lactate dehydrogenase in the screened strain
Lactate-producing characteristics: After activation with MRS seed medium, the selected strain was inoculated at a ratio of 5% (v/v) into the fermentation medium and cultured at 50 °C in a shaker at a shaking speed of 180–200 rpm/min for 48 h. Meanwhile, the growth, pH change and lactate production of the strain were determined during different growth periods. The lactate assay was performed according to the manufacturer’s instructions [Citation16].qRT-PCR was performed to determine the relative expression of lactate dehydrogenase genes LdhL1 and LdhL2 in the selected strain during various growth periods, and total RNA extraction, expression value determination and analysis of each sample were carried out with reference to the method of Lin et al. [Citation17]. Among them, primer sequences were listed as follows: LdhL1-F: 5′-CCATGCCGGGGCTTGCAAAAC-3′; LdhL1-R: 5′-GGCGCCGGGATTTTTT TCAA-3′; LdhL2-F: 5′-CTCTGCAATAGTATGACTTT-3′; LdhL2-R: 5′-CCTTTCAGGTCAAGCCCGA-3′; 16S rRNA (internal reference gene)-F: 5′-GCATGGAGGAAAAAGGAA-3′; 16S rRNA (internal reference gene)-R: 5′-TAAAACTCTGTTGCCGGG-3′. The measured value CT ≥2 was used as the criterion for high expression [Citation18].
Results
Isolation and identification of strains
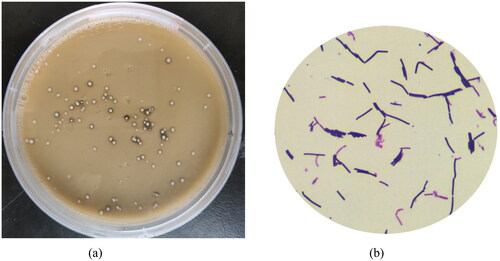
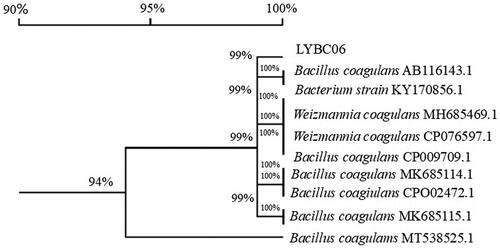
The colonies of the strains isolated from the digestive tract mucosa of fattening pigs were grown on MRS medium as gray-white, round, convex, moist, shiny and neat-edged colonies, which produced a transparent circle of decomposed calcium carbonate (). Under microscope observation, the strain was rod-shaped, blunt and round at both ends, with oval spores, terminal growth, no flagella and Gram-positive reaction (). The PCR product size of the isolated strain was about 1400 bp. The primer sequence obtained was compared by NCBI Blast to construct NJ evolutionary tree. The sequence homology between the isolated strain and the strain with downloaded sequence information was more than 94%. Therefore, the obtained strain belongs to Bacillus coagulans (), named LYBC06. After identification, the strain was submitted to China General Microorganism Culture Collection and Management Center for preservation, and the preservation number was CGMCC1.19347.
Analysis of strain resequencing results
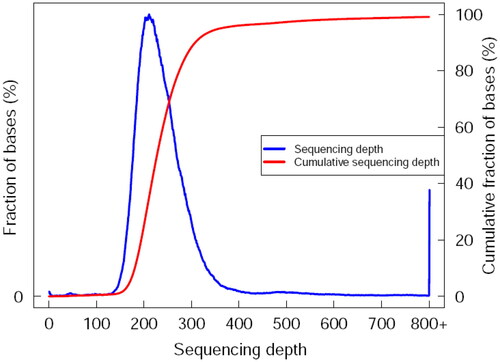
Alignment results to the reference genome. The raw base of raw sequencing data obtained by high throughput sequencing technology (NGS) for LYBC06 strain was 1058777700 bp, with an effective rate of 99.70% and GC content of 46.14%. Efficient sequencing data were aligned to the reference genome using the BWA program, and the results were sorted by SAMtools with a mapping rate of 80.41%, coverage at 1× at least 81.01% and coverage at 4X at least 80.84%.The distribution and cumulative distribution of sequencing depth for LYBC06 strain was shown in , and the data obtained from the sequencing of the LYBC06 strain revealed a good coverage with the reference genome.
Figure 3. Sequencing depth and cumulative distribution of LYBC06 strain. Note: The data obtained from the sequencing of the LYBC06 strains revealed a good coverage with the reference genome, the sequencing depth coverage at least 1× was 81.01%, and coverage at least 4× was 80.84%.
ANNOVAR software was used to perform a variant annotation of the genetic variation information for LYBC06 strain through SNP, InDel, SV and CNV. Compared with the reference sequence, the genome-wide heterozygous ratio of the LYBC06 strain was 0.042‰, with 14645 total variant SNP sites (0.4273% of the whole genome), 625 total InDel mutations (0.0182%), 106 total CNV due to gain and loss (0.0031%), and 285 total SV annotated variants (0.0083%). Information searching revealed that the mutated genes were mainly involved in hypothetic protein, glycogen/starch/alpha-glucan phosphorylase, spore protein, serine/threonine protein kinase, glycerol-3-phosphate acyltransferase, l-lactate dehydrogenase and other genes related to strain growth, regulation of energy metabolism, protease and lactate production, which were distributed on several sequences of chromosome NZCP009709.1.
Three-generation whole-genome sequencing of strain and functional annotation analysis
The high-quality genome sequence of LYBC06 strain was obtained through the PacBioRS II sequencing platform, the reads were assembled with the software SMRT Link version 5.0.1, and genome assembly was optimized by the arrow software, thus resulting in a final genome size of 3385106 bp and GC content of 46.86%. The genome prediction using GeneMarkSVersion 4.17 resulted in 4092 coding genes with an average length of 711 bp, and the total length of coding regions accounted for 85.96% of the whole genome. An additional 15 non-coding RNAs were obtained. A total of 36 gene islands (GIs) with an average length of 11735 bp and 7 prophages with an average length of 45140.3 bp were also obtained. The raw data obtained from the sequencing were uploaded to the NCBI database with BioProject ID: PRJNA752963.
The predicted protein-encoding genes from LYBC06 strain were input into the GO, KEGG, COG, NR, Pfam, TCDB and Swiss-Prot databases for alignment, and the number of annotated genes (non-redundant) in each database was 2297 for GO, 3312 for KEGG, 2308 for COG, 3726 for NR, 2297 for Pfam, 317 for TCDB and 1914 for Swiss-Prot, respectively. Among them, the most numerous metabolic pathway categories in KEGG annotation were: global and overview maps, carbohydrate metabolism, amino acid metabolism, metabolism of cofactors and vitamins, membrane transport and energy metabolism (). Among the COG annotation, the most numerous category of metabolic pathways was amino acid transport and metabolism, followed by carbohydrate transport and metabolism, as well as energy production and conversion (), suggesting that LYBC06 strain was involved in relatively-active amino acid, carbohydrate and energy metabolism. A total of 109 resistance genes were annotated in the CARD database (The Comprehensive Antibiotic Research Database), and the top 13 resistance genes with the largest number of genes were shown in . The top 3 resistance genes with the largest number of genes were ARO: 3000535 (22), ARO: 3002987 (11), and ARO: 3003980 (8), associated with macrolide resistance, bacitracin resistance and tetracycline resistance genes, respectively.
Figure 4. KEGG analysis and statistics of LYBC06 stain. Note: The predicted protein-encoding genes of LYBC06 strains in KEGG annotation, the most numerous metabolic pathway categories were: global and overview maps, carbohydrate metabolism, amino acid metabolism, metabolism of cofactors and vitamins, membrane transport and energy metabolism.
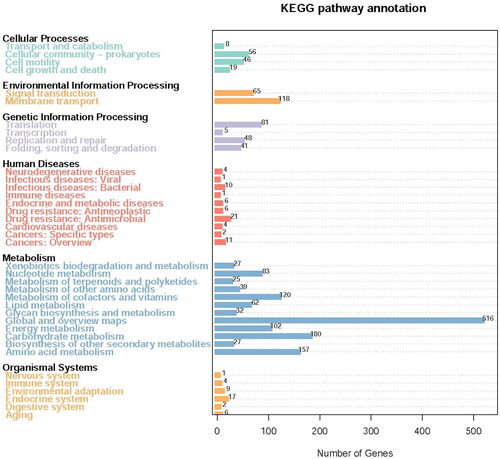
Figure 5. COG function classification of LYBC06 stain. Note: The predicted protein-encoding genes of LYBC06 strains in COG annotation, the most numerous category of metabolic pathways was amino acid transport and metabolism, followed by carbohydrate transport and metabolism and energy production and conversion.
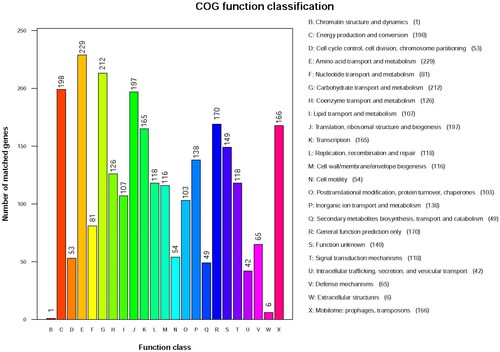
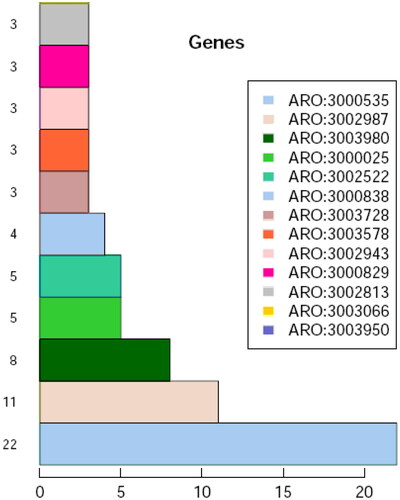
Figure 6. Number of notes in CARD database of the first 11 resistance genes of LYBC06 strain. Note: The predicted protein-encoding genes of LYBC06 strains in CARD database annotation, the top 3 resistance genes with the largest number of genes were ARO: 3000535 (22), ARO: 3002987 (11), and ARO: 3003980 (8).
The mapping of genome circles for LYBC06 strain was shown in , and the complete genomic sequence was finally obtained through whole genome sequencing. A circular chromosome with a full length of 3.38 MB was also obtained. After searching its whole genome information, the statistical information of lactate dehydrogenase genes was shown in , including four lactate dehydrogenase genes in LYBC06 strain, three of which were related to the production of l-lactate, such as 3 LdhL1 genes and 1 LdhL2 gene.
Figure 7. Genome circle of LYBC06 strain. Note: The whole genome sequence of LYBC06 strain was a circular chromosome with a total length of 3.38MB. From the outside to the inside, they were the location coordinates of the genome sequence, the encoded genes, the results of gene function annotation, ncRNA, the GC content of the genome and the GC skew value of the genome.
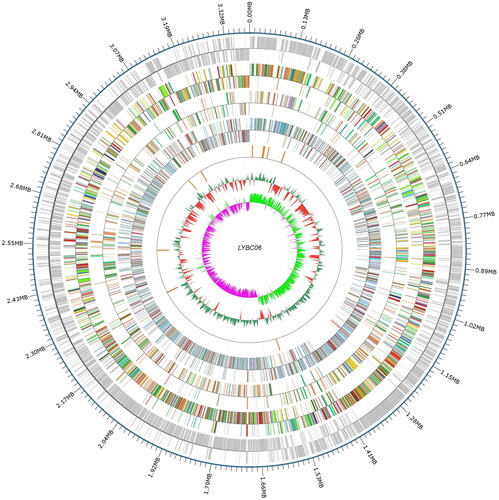
Table 1. Analysis of lactate dehydrogenase related genes in the genome of LYBC06 strain.
Comparative analysis for the genome of the strains
The genome information of three B. coagulans strains with homology over 99% was downloaded from the NCBI website, and the basic characteristics of the genome were shown in . The LYBC06 strain was relatively similar to other Bacillus coagulans from different origins based on genome size, GC content, protein-coding sequence, average CDS size, coding region occupancy and the number of RNA genes, indicating that the LYBC06 strain had typical characteristics of Bacillus coagulans. Comparison of all coding genes in LYBC06 strain with other three strains using BLASTp revealed that 2530 genes were common genes for all four strains and 809 genes was unique to LYBC06 strain (). Analysis for proteins encoded by these peculiar genes revealed that some proteins, such as transcription-regulating proteins, enzymes and unknown proteins contributed to the peculiar genetic properties of the LYBC06 strain. Collinearity analysis of four B. coagulans strains by Mauve software indicated that four strains had relatively good collinearity, with four locally collinear blocks (LCBs). A number of translocations and inversions, and a small number of genomic rearrangement events, such as inversions and deletions, were observed among the genomes of the four strains ().
Figure 8. Common genome of four Bacillus coagulans strains. Note: The strains had 2530 common protein coding genes, while LYBC06 train had 809 specific protein coding genes.
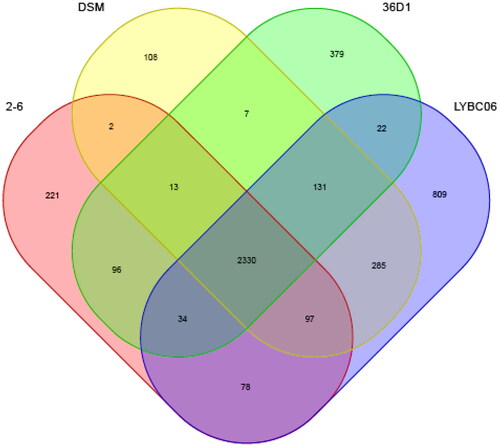
Figure 9. Synteny block of four Bacillus coagulans strains genomes. Note: The same color module represents the collinear region. Good co-linearity of the LYBC06 strain and other Bacillus coagulans strains and 4 locally co-linear blocks (LCBs) were shown.
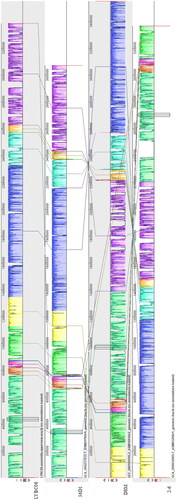
Table 2. General genomic information of 4 Bacillus coagulans strains.
Lactate-producing characteristics of the strain and gene expression level of lactate dehydrogenase
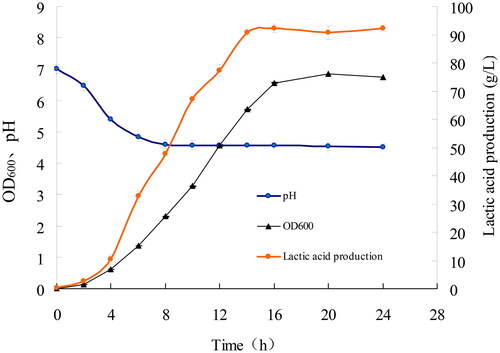
The growth, pH change and lactic acid production of LYBC06 strain in fermentation medium were shown in . The results showed that lactic acid production was basically synchronized with the growth of LYBC06 strain, with larger amount of bacterial growth and rapid production of lactic acid up to 92 g/L during the 4-16 h logarithmic growth phase. Subsequently, bacterial growth and lactate production reached to a stable level. In terms of pH change, during the early stage of bacterial growth (0–6 h), the pH revealed the rapid decrease and sequentially it was stabilized at pH 4.5. The major reason may be that calcium carbonate suspended in the initial fermentation broth was not enough to neutralize the produced lactic acid, thereby resulting in the rapid decrease in pH. The resultant lactic acid can be neutralized by the constant suspension of calcium carbonate deposited at the bottom, thereby keeping the stable pH subsequently.
Figure 10. Growth, pH change and Lactic acid production curve of LYBC06 strain. Note: The lactic acid production was basically synchronized with the growth of LYBC06 strains, and the maximum amount of lactate could be produced up to 92 g/L at 16 h culture during the logarithmic growth phase. In terms of pH change, during the early stage of bacterial growth (0–6 h), the pH revealed a rapid decrease and sequentially, it was stabilized at pH 4.5.
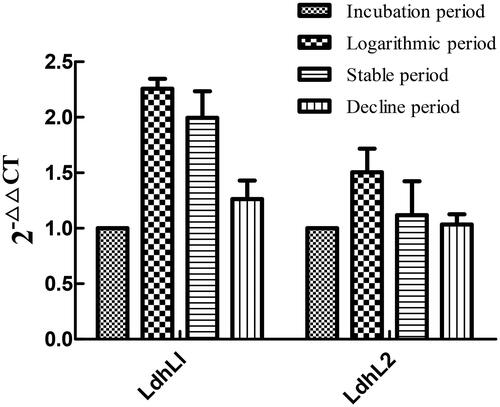
The 1% agarose gel electrophoresis test indicated that the quality of total RNA extract was high in all four samples for latency phase, logarithmic phase, stationary phase and decay phase of LYBC06 strain. The expression levels of LdhL1 and LdhL2 genes in four samples were analyzed by qRT-PCR, as shown in . The 2–△△CT values of LdhL1 and LdhL2 gene expression at the transcriptional level in LYBC06 strain during different growth periods all showed an initial increase followed by a sequential decrease, with the highest expression level at the logarithmic phase. The experimental determination results were consistent with the production of lactate in the previously mentioned strain. The relative expression level of LdhL1 gene was higher than that of LdhL2 gene during different growth periods. Among them, the 2–△△CT value of LdhL1 gene at the logarithmic phase was 2.26, and the relative expression value reached the overexpression level (≥2), indicating that LdhL1 gene plays the more important role in lactate production in LYBC06 strain.
Figure 11. Relative expression value of lactate dehydrogenase gene in LYBC06 strain at different growth stages. Note: The 2-△△CT values of LdhL1 and LdhL2 gene expression at the transcriptional level in LYBC06 strains during different growth periods all showed an initial increase followed by a sequential decrease, with the highest expression level at the logarithmic phase, and the relative expression level of LdhL1 gene was higher than that of LdhL2 gene during different growth periods.
Discussion
Recently, B. coagulans has gradually attracted attention as a new type of microbial preparation for feeding, and its characteristics of high temperature and stress resistance, as well as lactic acid production, offer it as a good candidate for replacing antibiotic applications in the feed industry. Practice has proved that feeding livestock and poultry with Bacillus coagulans can significantly increase the number of lactic acid bacteria in the intestinal tracts of livestock and poultry, reduce intestinal pH, reduce the odor of ammonia in the livestock house and also can increase the feed intake of livestock and poultry, reduce feeding costs, and promote daily body weight gain and a significant increase in feed conversion rate [Citation19,Citation20].
Re-sequencing of LYBC06 strain showed that its genome, especially coding genes, was less variable overall. However, the variants in functional genes encoding hypothalamic protein, glycogen/starch/alpha glucan phosphorylase, spore protease, serine/threonine protein kinase, glycerol-3-phosphate acyltransferase, l-lactate dehydrogenase and phenyl lactate dehydrogenase associated with the growth, regulation of energy metabolism, protease production and lactate production of the strain, were enhanced, indicating that the isolated LYBC06 strain has strong protease- and lactate-producing capacity. Whole genome sequencing and functional annotation of the protein-encoding genes revealed that LYBC06 strain was involved in a variety of metabolic pathways and metabolic functions, including the metabolism of carbohydrates, amino acids, cofactors and vitamins, membrane transport and energy. The higher number of genes, annotated with amino acid transport and metabolism, carbohydrate transport and metabolism, energy production and conversion, indicate that LYBC06 strain was involved in more active metabolism of amino acids and carbohydrates. The addition of LYBC06 strain into microecological formula is beneficial to promote the transformation and absorption of nutrients for livestock and poultry. Functional annotation of the CARD database indicated that the genome of the LYBC06 strain is strongly resistant to macrolides, peptides and tetracyclines, but some microbial strains have the problem with inconsistence with the genotype and phenotype of antibiotic resistance genes, so further validation is highly required for antibiotic phenotyping to provide the guidance for the preparation of LYBC06 strain and the combinatorial applications with antibiotics [Citation21]. Comparative genomic analysis showed that the basic characteristics of the genome in LYBC06 strain were more similar to other B. coagulans strains, with the typical characteristics of the species. Several B. coagulans stains have relatively good collinearity, but the LYBC06 strain still has 809 unique genes, leading to its unique genetic characteristics [Citation22]. The whole genome information of the LYBC06 strain clarifies the basic information of its lactate dehydrogenase genes. The lactate dehydrogenase genes in B. coagulans confirmed to be related to the production of l-type lactate are LdhL1 and LdhL2 genes [Citation23]. In this study, the lactate production assay and the expression analysis of LdhL1 and LdhL2 genes in LYBC06 strain at different growth periods showed an initial increase followed by a sequential decrease, with the highest expression level up to 92 g/L during the logarithmic growth phase, which is basically consistent with the growth of bacteria and the production of lactate, and the production of lactate in the LYBC06 strain was higher than that of other different sources of B. coagulans [Citation24]. LdhL1 gene played an important role in the production of lactate in LYBC06 strain, and the relative expression value of LdhL1 gene was higher than that of LdhL2 at different growth periods of LYBC06 strain, so we speculate that LdhL1 gene expression is an important reason for l-type lactic acid production in B. coagulans and can be used as an important phenotypic gene for the studies of this species.
With the continuous improvement of biotechnology, high-throughput sequencing [Citation25] and qRT-PCR [Citation26] techniques have become important strategies and analytic methods for the studies on microbial genomics and gene expression, which can provide important technical methods for the studies on gene structure, functional properties and functional gene associated with metabolism, regulation and interaction in microbial strains. In this study, the whole genome information of the B. coagulans strain was comprehensively mastered by resequencing, whole genome sequencing and transcriptional analysis of lactate dehydrogenase genes, which is beneficial to reveal the essence of gene compositions and functional specificity of B. coagulans at the molecular level and to provide a theoretical basis for its applications in bio-fermentation feeds, functional foods and biomedicine.
Conclusions
In this study, a B. coagulans LYBC06 strain with high lactate-producing capacity was isolated from mucosal tissue of the digestive tracts of fattening pigs. The overall variation in the genome, especially in the coding genes, was small in LYBC06 strain, but the function gene variants related to strain growth, regulation of energy metabolism and the production of proteases and lactate, involved in the metabolism of amino acids and carbohydrates, were relatively active and enhanced, suggesting the strong resistance to macrolides, peptides and tetracyclines. The whole genome information of the LYBC06 strain was obtained, which revealed good collinearity with several B. coagulans strains from other sources, with typical characteristics of the species. Bacterial growth, gene expression of lactate dehydrogenase and lactate production in LYBC06 strain were largely synchronized, and the maximum produced amount of lactate could be up to 92 g/L at 16 h culture during the logarithmic growth phase, when the LdhL1 gene played an important role in the production of lactate in LYBC06 strain.
Author contributions
BSL performed the investigation, analyzed the data and wrote the original draft. DYH, WYW and BL provided resources and input on the experimental design, methodology and data analyses. YL conceived and designed the study and acquired research funding. XHC revised and edited the manuscript and carried out formal data analysis. All authors read and approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings from this study are available from the corresponding authors [YL and XHC] upon reasonable request.
Additional information
Funding
References
- Shi S, Liu J, Dong J, et al. Research progress on the regulation mechanism of probiotics on the microecological flora of infected intestines in livestock and poultry. Lett Appl Microbiol. 2022;74(5):647–655.
- Kumar S, Tariq H, Singh A, et al. Significance of probiotics as feed additives in livestock and poultry nutrition. Indus J Anim Nutr. 2017;34(4):361–373.
- Midhun SJ, Neethu S, Vysakh A, et al. Antagonism against fish pathogens by cellular components/preparations of Bacillus coagulans (MTCC-9872) and it’s in vitro probiotic characterisation. Curr Microbiol. 2018;75(9):1174–1181.
- Cubas-Cano E, Venus J, González-Fernández C, et al. Assessment of different Bacillus coagulans strains for l-lactic acid production from defined media and gardening hydrolysates: effect of lignocellulosic inhibitors. J Biotechnol. 2020;323:9–16.
- Zhang B, Zhang H, Yu Y, et al. Effects of Bacillus coagulans on growth performance, antioxidant capacity, immunity function and gut health in broilers. Poult Sci. 2021;100(6):e101168.
- Amoah K, Huang QC, Tan BP, et al. Dietary supplementation of probiotic bacteria, Bacillus coagulans ATCC 7050, improves the growth performance, intestinal morphology, microflora, immune response, and disease confrontation of pacific white shrimp, Litopenaeus vannamei. Fish Shellfish Immunol. 2019;87:796–808.
- Bucher T, Keren-Paz A, Hausser J, et al. An active β-lactamase is a part of an orchestrated cell wall stress resistance network of Bacillus subtilis and related rhizosphere species. Environ Microbiol. 2019;21(3):1068–1085.
- Rosser CL, Jin L, Beauchemin KA, et al. 0471 The effect of binding feed enzymes to spores of Bacillius subtlis and Bacillius coagulans on in vitro NDF digestibility in ruminal batch cultures. J Anim Sci. 2016;94(suppl_5):e225–225.
- Chauhan D, Patel Y, Doshi JA, et al. Draft genome sequence of Bacillus coagulans ZB29, a commercial probiotic strain. Microbiol Resour Ann. 2019;8:e01125.
- Fang Q, Shu JJ, Tao L, et al. Isolation and identification of thermostable lactase-producing Bacillus coagulans T242. Adv Mat Res. 2013; 634–638:1253–1258.
- Adeoti OM, Usman AT. The molecular characterization of Rhizobacteria isolates from Saki, Nigeria. EJBIO. 2021;2(2):26–38.
- Rong M, Zheng X, Ye M, et al. Phenotypic plasticity of Staphylococcus aureus in liquid medium containing vancomycin. Front Microbiol. 2019;10:e809.
- Green MR, Sambrook J. Molecular cloning: a laboratory manual. 4th ed. New York: Cold Spring Harbor Lab Press; 2012; p. 20–25.
- Lin BS, Song ZZ, Jia YL, et al. Biological characteristics and genome-wide sequence analysis of endophytic nitrogen-fixing bacteria Klebsiella variicola GN02. Biotechnol Biotechnol Equip. 2019;33(1):108–117.
- Darling AE, Mau B, Perna NT. ProgressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5(6):e11147.
- Luo J, Lin BS, He YQ, et al. Comparative analysis of analytical methods of lactic acid content in microbial fermentation feeds. Feed Rev. 2012;5:37–39 (in Chinese).
- Lin BS, Liu JM, Zhang X, et al. The flora compositions of nitrogen-fixing bacteria and the differential expression of nifH gene in Pennisetum giganteum z.x.lin roots. BioMed Res Int. 2021;2021:1–7.
- Li Y, Liu F. C. Analysis of the copy number of exogenous genes in transgenic cotton using real-time quantitative PCR and the 2-△△CT method. Afr J Biotechnol. 2012;11:6226–6233.
- Hung AT, Lin SY, Yang TY, et al. Effects of Bacillus coagulans ATCC 7050 on growth performance, intestinal morphology, and microflora composition in broiler chickens. Anim Prod Sci. 2012;52(9):874–879.
- Gu SB, Zhao LN, Wu Y, et al. Potential probiotic attributes of a new strain of Bacillus coagulans CGMCC 9951 isolated from healthy piglet feces. World J Microbiol Biotechnol. 2015;31(6):851–863.
- Yi Z, Ding QJ, Wang HC, et al. Genetic diversity and biological characteristics of Clostridium butyricum based on comparative genomics. Food Ferment Indus. 2020;46:5–12 (in Chinese).
- Sun L, Zhang C, Lyu P, et al. Contributory roles of two l-lactate dehydrogenases for l-lactic acid production in thermotolerant Bacillus coagulans. Sci Rep. 2016;6:e37916.
- Wang L, Cai Y, Zhu L, et al. Major role of NAD-dependent lactate dehydrogenases in the production of l-lactic acid with high optical purity by the thermophile Bacillus coagulans. Appl Environ Microbiol. 2014;80(23):7134–7141.
- Coelho LF, Beitel SM, Sass DC, et al. High-titer and productivity of l-(+)-lactic acid using exponential fed-batch fermentation with Bacillus coagulans arr4, a new thermotolerant bacterial strain. J Biotech. 2018;8:e213.
- Li J, Sang C, Yang J, et al. Stoichiometric imbalance and microbial community regulate microbial elements use efficiencies under nitrogen addition. Soil Biol Biochem. 2021;156:e108207.
- Wang XY, Peng F, Dong G, et al. Identification and validation of appropriate reference genes for qRT-PCR analysis in Corynebacterium glutamicum. FEMS Microbiol Lett. 2018;365:1–8.