Abstract
Rice koji is a raw material used in the production of sake; however, details regarding the microbial flora and their dynamics in rice koji during the production process are poorly understood. Clarifying these issues can contribute to proposing a method and evaluation that will improve the quality of rice koji and sake. The aim of this study was to determine the microflora in rice koji and the effectiveness of the traditional microbial control techniques used in the sake production process. We analyzed the diversity and changes in bacterial flora during rice koji production by amplicon sequencing of the 16S rRNA gene. The predominant taxon in all rice koji samples was family Staphylococcaceae. The microbial population and the changes in its distribution for five consecutive stages in rice koji production were examined by direct colony counting. Bacteria counts in all samples were below the limit of detection initially, then increased rapidly toward the final stage. The predominant bacterial colonies from all samples were yellow and were identified as Staphylococcus gallinarum through 16S rRNA gene sequence similarity. The S. gallinarum isolates exhibited faster growth in pregelatinized rice medium. Interestingly, the growth of S. gallinarum isolates was suppressed by low temperature (12 °C), ethanol concentration (≥6%) and the addition of lactic acid, which are traditional microbial control methods used during sake fermentation. Therefore, proper control of the traditional sake production process can effectively inhibit the growth of undesirable bacteria such as S. gallinarum that emerge during the production of rice koji.
Introduction
Sake is a traditional alcoholic beverage made in Japan. The raw materials used in sake production are mainly rice, rice koji [Citation1] and water. Briefly, the rice koji breaks down the rice starch into glucose, and the glucose is simultaneously fermented in the sake mash, a process referred to as parallel double fermentation. Finally, the alcohol concentration of sake reaches around 18%, which is the highest concentration for a brewed alcoholic beverage. Generally, these fermentation processes are conducted in open tanks, and the prevention of microbial contamination in sake brewing factories remains an important issue. Traditional techniques have been developed and are still used as measures to prevent microbial contamination during sake production. To prevent wild yeast contamination, pure culture of brewing yeast is added in the early phase of fermentation, and with the subsequent yeast growth, the raw materials are added in three time points. Another way to prevent contamination from wild yeast and undesirable bacteria is to incorporate lactic acid bacteria into the sake mash (i.e. the traditional technique referred to as Kimoto or Yamahai-moto) or to use the modern method of lactic acid treatment (referred to as Sokujo-moto). In addition, the growth of microorganisms other than fermenting yeast in the mash is inhibited by keeping the temperature at a low level (6–12 °C) and by a gradual increase in the concentration of alcohol (up to 18% approx.) produced by fermenting yeast. High alcohol concentration in sake is a traditional spontaneous condition which is known as a factor for microbial suppression. However, even using these methods, bacterial contamination can still occur during sake production. Although uncommon, the worst problem is caused by alcohol-resistant lactic acid bacteria, which lead to an acidic taste, a strange odor and cloudiness in the sake or the sake mash (referred to as Hiochi) [Citation2]. The sources of the contaminating microorganisms are unclear, but the contamination is thought to occur during the brewing process.
During the production of rice koji, steamed rice containing Aspergillus oryzae is maintained under optimal conditions for microbial growth, which supports the growth of not only A. oryzae, but also undesirable microorganisms. The production of rice koji takes approximately 2 days and consists of first steaming the rice at nearly 100 °C and then letting it cool to 20–25 °C, followed by the addition of A. oryzae spore powder. This process involves several steps, including manually mixing and inverting the top and bottom layers of the rice mass. Thus, the risk for microbiological contamination is higher during rice koji production than in the subsequent fermentation process. Undesirable microorganisms produce compounds such as 2,4,6-trichloroanisole and guaiacol that result in off-flavors, thereby reducing the quality of the sake [Citation3].
Microorganisms growing on rice koji at the stage of alcoholic fermentation have been reported by various researchers using conventional dilution plating techniques [Citation3, Citation4] and molecular identification methods [Citation5–7]. Microorganisms present during the early stages of alcoholic fermentation in sake production are likely to be derived from the rice koji. Previous studies reported the presence of Bacillus spp. and Staphylococcus spp. in the early stages of sake fermentation [Citation5, Citation8]. These bacteria are considered to be the most common microorganisms that produce guaiacol, a potent phenolic off-odor compound, in the sake brewing process. Staphylococcus spp. have not been isolated in the middle and late stages of sake fermentation, and it remains unclear why the presence or absence of these bacteria depends on the duration of the fermentation process.
The present study aimed to reveal the distribution of microorganisms during rice koji production and demonstrate how to suppress contamination from undesirable microorganisms in a simulated sake production environment using traditional microbial control techniques.
Materials and methods
Rice koji samples
The seven rice koji samples (Supplemental Table S1) used in the present study were supplied by three different sake breweries located in Yamanashi Prefecture, Japan, from 2019 to 2022. These three breweries are located at least 10 km away from each other. They also use different lots of raw materials while producing rice koji, and their workers do not interact with each other. Each sample was collected from the mixed rice koji at 3 or 4 equal points with food sanitation gloves and was immediately packed in a polyethylene bag and was stored in a freezer at −30 °C until use.
Amplicon sequencing
All rice koji samples were lyophilized using a VD-250R Freeze Dryer (Taitec Corporation, Saitama, Japan). For DNA extraction, Lysis Solution F (Nippon Gene Co., Ltd., Tokyo, Japan) was added to the crushed samples and allowed to stand at 65 °C for 10 min. The supernatant was subsequently centrifuged at 12,000 g for 2 min and then aliquoted. DNA was purified from the aliquoted supernatant using the MPure-12 System and the MPure Bacterial DNA Extraction Kit (MP Bio). Extracted DNA was tested for quality and quantity via BioTek Synergy LX (Agilent, Santa Clara, CA, USA) and QuantiFluor dsDNA System (Promega Corporation, Madison, WI, USA). Libraries were prepared using a two-step tailed polymerase chain reaction (PCR) method targeted for V3-V4 region of 16S rRNA gene with anti-plastid peptide nucleic acid (PNA) and anti-mitochondrial PNA, and sequenced at 2 × 300 bp using the MiSeq system and MiSeq Reagent Kit v3 (Illumina). The conditions for the 1st PCR were as follows: the primers, 1st-341f_MIX (forward) (5ʹ-ACACTCTTTCCCTACACGACGCTCTTCCGATCT-NNNNN-CCTACGGGNGGCWGCAG-3ʹ), 1st-805r_MIX (reverse) (5ʹ-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT-NNNNN-GACTACHVGGGTATCTAATCC-3ʹ), anti-plastid PNA (5ʹ-GGCTCAACCCTGGACAG-3ʹ) and anti-mitochondrial PNA (5ʹ-GGCAAGTGTTCTTCGGA-3ʹ); reaction mixture, 2 × PCR buffer for KOD FX Neo 5.0 μL, dNTPs (each 2 mmol/L) 2.0 μL, 10 μmol/L forward primer 0.5 μL, 10 μmol/L reverse primer 0.5 μL, 100 μmol/L anti-plastid PNA 0.375 μL, 100 μmol/L anti-mitochondrial PNA 0.375 μL, template DNA 1.0 μL, KOD FX Neo (1.0 U/μL) 0.2 μL; PCR program, 94 °C 2 min, 30 cycles (98 °C 10 s, 55 °C 30 s, 68 °C 30 s), 68 °C 7 min. The conditions for the 2nd PCR were as follows: primers, 2ndF (forward)(5ʹ-AATGATACGGCGACCACCGAGATCTACAC-Index2-ACACTCTTTCCCTACACGACGC-3ʹ) and 2ndR (reverse) (5ʹ-CAAGCAGAAGACGGCATACGAGAT-Index1-GTGACTGG-AGTTCAGACGTGTG-3ʹ); reaction mixture, 10x Ex Buffer 1.0 μL, dNTPs (each 2. 5 mmol/L) 0.8 μL, 10 μmol/L forward primer 0.5 μL, 10 μmol/L reverse primer 0.5 μL, 1st PCR product 2.0 μL, ExTaq HS [TaKaRa] (5 U/μL) 0.1 μL; PCR program, 94 °C 2 min, 12 cycles (94 °C 30 s, 60 °C 30 s, 72 °C 30 s), 72 °C 5 min. The sequencing was performed by Bioengineering Lab. Co., Ltd. (Sagamihara, Japan). The sequencing reads were curated and the taxonomic diversity was determined using the EzBioCloud Microbiome Taxonomic Profile (MTP) pipeline with the PKSSU4.0 database. The sequence data obtained in this study have been deposited in the Sequence Read Archive (DRA) of the DNA Data Bank of Japan (DDBJ) under accession number PRJDB15050.
Physicochemical analysis of rice koji
Two rice koji samples from 2019 (one sample each from the SA-1 production process series and SB-3 production process series) supplied from different sake breweries located in Yamanashi Prefecture, Japan, were used for the physicochemical analyses. The moisture content of rice koji was measured using MOC-102H moisture analyzer (Shimadzu Corporation, Kyoto, Japan) and is indicated as grams per 100 grams of rice koji. The homogenized rice koji and distilled water were mixed in equal amounts, and the sugar content and composition of glucose in the supernatant were measured using a DBX-55 digital refractometer (Atago Co., Ltd., Tokyo, Japan) and Prominence modular HPLC (detector; RID-10A refractive index detector, pump; LC-10ADVP, column oven; CTO-10AVP, Shimadzu Corporation), respectively. The HPLC column was SCR-101C (300 × 7.9 mm with Ca type cation exchanger consisting of styrene-divinylbenzene copolymer, Shimadzu Corporation). Ultrapure water was used as the mobile phase. The flow rate was 0.8 mL/min. The column temperature was 80 °C. The injection volume was 10 μL. Enzyme activities of α-amylase and glucoamylase were measured using test kits (Kikkoman Biochemifa Co., Tokyo, Japan) according to the manufacturer’s instructions.
Assessment of changes in bacterial population during rice koji production
To isolate the various microorganisms, approximately 10 g of rice koji collected from each processing stage supplied from brewery A and B was suspended in 10 mL of sterile saline solution and diluted 10 times. The resulting samples were plated on potato dextrose agar "Nissui" (PDA, 05709, Nissui Pharmaceutical Co., Ltd., Tokyo, Japan) for isolation of yeast or mold, and standard assay agar "Nissui" (05618, Nissui Pharmaceutical Co., Ltd.) for isolation of bacteria. PDA and standard assay agar plates were cultured at 25 °C for 7 days and 35 °C for 2 days, respectively. Five plates for each sample in each medium were used for colony counting.
Identification of bacterial isolates
Bacterial colonies were grouped by their colors and were randomly picked up. All bacterial colonies were purified on standard assay agar plate, and their 16S rRNA gene sequences were determined to clarify their taxonomic positions. DNA extraction and 16S rRNA gene sequencing were conducted according to the method described previously [Citation9]. Briefly, cultured bacterial isolates were subjected to DNA extraction using PrepMan™ Ultra reagents (4318930, Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. PCR amplification of the 16S rRNA gene was carried out following the procedure described previously [Citation10] and directly sequenced by a commercial sequencing service (FASMAC, Kanagawa, Japan). The partially complete 16S rRNA gene sequences were compared to the published sequences in EzBioCloud (www.ezbiocloud.net). A 16S rRNA gene sequence similarity value of greater than 98.7% could be assigned to the genus/species level.
Phylogenetic analysis of isolates
For phylogenetic analysis of isolates identified as S. gallinarum, we obtained reference sequences from the European Molecular Biology Laboratory/GenBank/DDBJ databases and aligned them using CLUSTAL_X. Phylogenetic trees were constructed by the neighbor-joining method using Molecular Evolutionary Genetics Analysis software, version 10.1.8. The topologies of the constructed trees were evaluated by bootstrap analysis with 1,000 resamplings.
Growth of S. gallinarum isolates in pregelatinized rice
Pregelatinized rice AA-70 (Tokushima Koji Producing Co., Ltd., Tokushima, Japan) was used as culture medium to observe the behavior of S. gallinarum as a simulation of the initial process of rice koji production. The pregelatinized rice was sterilized at 110 °C for 5 min, and then 15 g of the rice was mixed with 4.5 mL of sterile water. Bacillus subtilis JCM 1465T and Staphylococcus aureus JCM 20624T were purchased from JCM (RIKEN BioResource Research Center, Ibaraki, Japan) as controls. Broth cultures of S. gallinarum strains A1 and A2, Bacillus subtilis JCM 1465T and Staphylococcus aureus JCM 20624T were adjusted to optical density (OD660) = 0.05 with 1,000-times dilution, then incubated at 38 °C. The number of bacterial colonies was counted on a BD Bacto SCD agar plate (Thermo Fisher Scientific) using the dilution plating method at 0, 1, 3 and 6 h.
Growth temperature, alcohol tolerance and lactic acid sensitivity of S. gallinarum isolates
The growth temperature, alcohol tolerance and lactic acid sensitivity of S. gallinarum isolates were evaluated using rice koji extract as a brewing simulation of sake mash. Rice koji extract was prepared from dried rice koji 1-65 (Tokushima Koji Producing Co., Ltd.) with 4 times water by weight and processed at 55 °C for 6 h. The filtrate was autoclaved (121 °C 15 min) and used. Rice koji extract composition values were 16.9 Bx measured using a DBX-55 digital refractometer (Atago Co., Ltd.), 0.7 acid value measured by titration volume of 0.1 mol/L NaOH (196-02195, FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan), and pH 5.04 measured using a LAQUA F-72 pH meter (Horiba, Ltd., Kyoto, Japan). Since typical sake mash composition values were 12.7-14.5 Bx and 0.7-1.0 acid value, rice koji extract was determined to be usable for brewing simulation of sake mash.
S. gallinarum isolates were inoculated in 5 mL rice koji extract in 10 mL test tube at OD660 = 0.01 and incubated at 12 °C or 32 °C, then the absorbance was measured at 0, 1, 2, 3, 4.5, 7.5, 10.5, 13.5, 16.5, 19.5, 22.5, 25.5, 28.5, 38.5, 51.5, 72.5, 87.5, 102.5, 111.5 h.
Data analysis
Growth, growth temperature, alcohol tolerance and lactic acid sensitivity of S. gallinarum isolates were examined in three independent experiments, and values are shown as means with standard deviation (±SD). Data were analyzed using Microsoft Excel version 2019 (Microsoft Corporation, Redmond, WA, USA).
Results
Analysis of the microbial flora of rice koji
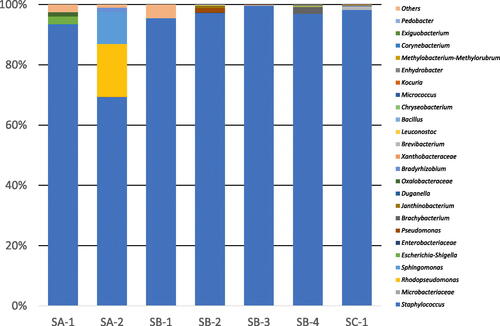
The microbial flora of seven rice koji samples were analyzed by amplicon sequencing of the V3/V4 region of the 16S rRNA gene. The results of the population analysis and distribution of observed taxa are shown in . The predominant taxon (65–95% abundance) in all samples of the rice koji belonged to the family Staphylococcaceae. The families Xanthobacteraceae and Microbacteriaceae were detected as minor representatives in all samples. The results of the population analysis and distribution of observed taxa at the genus level are shown in . The predominant bacteria were Staphylococcus spp.
Figure 1. Bacterial flora of rice koji collected from several breweries.
Notes: Letters (SA to SC) indicate the company and the numbers (1 to 4) indicate the sample number. Results for chloroplasts and mitochondria in rice koji were excluded. In samples SA-1 to SC-1, Staphylococcaceae spp. were predominant.
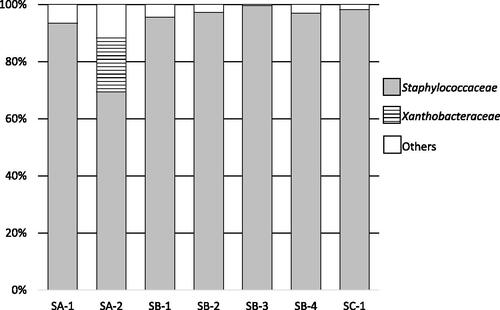
Figure 2. Bacterial flora of rice koji collected from several breweries at the genus level.
Notes: Letters (SA to SD) indicate the company and the numbers (1 to 4) indicate the sample number. Results for chloroplasts and mitochondria in rice koji were excluded. In samples SA-1 to SC-1, Staphylococcus were predominant.
Microbial population and its distribution in rice koji during each production stage
Rice koji is produced using a five-stage process. The temperature and time for microbial growth were recorded for each rice koji sample throughout the entire production process (Supplemental Figure S1 and Table S2). shows the number of microorganisms measured in the production of the SA-1 rice koji sample from brewery A. Bacterial colonies were not detected at 0 min (Tokomomi; see Supplemental Figure S1 for a description of the production process names), but then gradually increased to 2.7 × 105 colony-forming units (CFU)/g at 3060 min, i.e. 51.0 h (Dekoji). Yeast colonies were sparsely detected at 1740 min, i.e. 29.0 h (Mori), to 1970 min, i.e. 32.8 h (Shimai-Shigoto), and were not detected at 3060 min, i.e. 51.0 h (Dekoji). A. oryzae was detected at 9.7 × 103 CFU/g at 0 min (Tokomomi), a level that decreased transiently to 3.3 × 103 cells/g at 1740 min, i.e. 29.0 h (Mori), and increased at 3060 min, i.e. 51.0 h (Dekoji).
Figure 3. Changes in microbial profile during the five-stage koji production process for sample SA-1 from brewery A (a) and sample SB-3 from brewery B (b). an example of diagram for the mode of sake production process (rice koji production process and fermentation process) time-temperature (c).
Notes: Samples were collected at the end of each stage (Tokomomi, Kirikaeshi, Mori, Shimai-Shigoto, Dekoji) and the time was counted from the end of Tokomomi. Supplemental Figure S1 for a description of the stages. Filled triangles indicate the bacteria that were visually identified on standard assay agar. Filled circles indicate Aspergillus oryzae and filled squares indicate yeast; both were visually identified on potato dextrose agar (PDA). Colony-forming units (CFU) are presented as means of five replicate plates and standard deviations are shown as error bars. The vertical axis in (a) and (b) shows CFU in logarithmic scale. The data shown in (c) were taken from Tables S2 and S3, which were based on representative data.
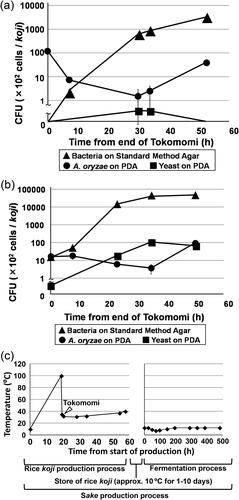
shows the number of microorganisms in the production of the SB-3 rice koji sample from brewery B. Bacteria were detected at a level of 1.2 × 103 CFU/g in the koji sample at 0 min (Tokomomi), and then increased rapidly from 4.1 × 103 CFU/g at 445 min, i.e. 7.4 h (Kirikaeshi), to 3.8 × 106 cells/g at 2935 min, i.e. 48.9 h (Dekoji). Yeast colonies were detected at 8.5 × 103 CFU/g, a level lower than the level of bacteria at all stages. A. oryzae was detected at a level of 102 CFU/g at each stage of the process.
shows the representative example of the mode of rice koji and sake production time-temperature diagram. The data were taken from Tables S2 and S3. The temperature of a rice koji production process was controlled at around 30 °C for 40 h after Tokomomi.
Until now, no major reasons have been found why the SA-2 minor population differs from the other samples. More detailed investigation of the source environment is needed.
Identification of bacterial isolates
The 16S rRNA gene of each isolate was amplified by PCR and sequenced by direct sequencing, and the identification results based on EzBioCloud similarity search are shown in . All isolates that showed 99.0% similarity or higher to known species were assigned tentatively with the name of the highest similarity taxon. The most frequently detected bacterium observed in most processing stages was Staphylococcus gallinarum except for 0 h in SB-3. The phylogenetic tree (Supplemental Figure S2) revealed that most isolates presumptively identified as S. gallinarum were classified as a single species. In all rice koji samples, the level of S. gallinarum remained high during the course of the production process. These data provided more accurate taxonomic information that the bacterium belongs to the family Staphylococcaceae as determined by the results of amplicon sequencing of the 16S rRNA gene (specifically the V3/V4 region).
Table 1. Identification of bacterial isolates based on 16S rRNA gene sequences from each rice koji processing stage.
Growth of S. gallinarum isolates in simulated rice koji
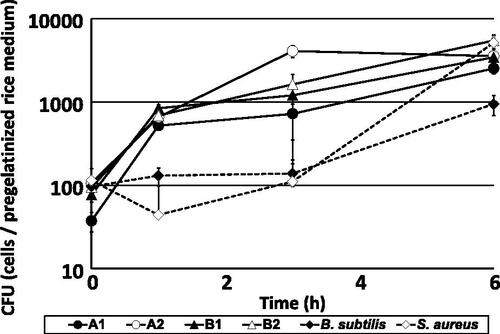
shows the number of CFU of representative S. gallinarum isolates obtained from breweries A and B, which were inoculated into pregelatinized rice medium. Compared to control bacteria, S. gallinarum isolates grew immediately within 1 h of incubation, then grew slowly over the subsequent 6 h. In contrast, the two control isolates showed rapid growth after 3 h of inoculation. These results suggest that the isolated S. gallinarum grows faster and tends to predominate in pregelatinized rice, which is the basis of rice koji and therefore used as a simulation of rice koji in the present study.
Figure 4. Changes in microbial counts of Staphylococcus gallinarum isolates and control strains in pregelatinized rice medium at 38 °C.
Notes: Isolates A1 (closed circle) and A2 (open circle) were obtained from brewery A, and isolates B1 (closed triangle) and B2 (open triangle) were obtained from brewery B. These isolates were already identified as S. gallinarum by 16S rRNA gene sequencing. Strains Bacillus subtilis JCM 1465T (closed rhombus) and S. aureus JCM 20624T were used as controls. Colony-forming units (CFU) are presented as means of five replicate plates, and standard deviations are shown as error bars.
Growth of S. gallinarum under traditional microbial control techniques
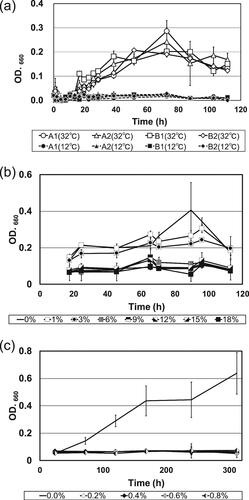
Generally, relatively low temperatures are used in the sake brewing process, and the alcohol concentration in the mash increases due to alcoholic fermentation. In addition, lactic acid is added to prevent the growth of bacteria [Citation4, Citation8]. shows the growth curves under different culture conditions of representative S. gallinarum isolates obtained from breweries A and B, which were inoculated into koji extract as a simulation of the brewing process. shows that none of the isolates exhibited any growth at 12 °C, which is usually the highest temperature of sake mash ( and Supplemental Table S3). In contrast, significant growth was observed at 32 °C as compared to 12 °C. shows the alcohol tolerance of isolates under alcohol concentrations of 1 to 18%. The highest concentration was selected based on the final alcohol concentration of sake being at least 18%. Isolates did not grow at alcohol concentrations greater than 6%. shows the lactic acid sensitivity of isolates under alcohol concentrations of 0 to 0.8%. The highest concentration of lactic acid was selected based on the highest concentration in sake mash used by breweries being at least 0.6%. Isolates did not grow at lactic acid concentrations greater than 0.2%. Thus, the isolates identified as S. gallinarum are unlikely to cause problems with healthy sake mash under typical brewing conditions. However, under unusual brewing conditions, such as a temperature of 32 °C or a lower alcohol concentration, these isolates could pose the risk of spoilage.
Figure 5. Growth curves of Staphylococcus gallinarum isolates in koji extract cultured at different temperatures, alcohol concentrations and lactic acid concentrations.
Notes: (a) Growth curves of S. gallinarum isolates cultured in koji extract medium at 12 °C and 32 °C, respectively. Strains A1 and A2 were obtained from brewery A, and strains B1 and B2 were obtained from brewery B. All data are presented as means of three replicate tubes, and standard deviations are shown as error bars. (b) Growth curves of S. gallinarum strain A2 cultured in koji extract medium containing various ethanol concentrations (0–18%, v/v). All data are presented as means of three replicate tubes, and standard deviations are shown as error bars. (c) Growth curves of S. gallinarum strain A2 cultured in koji extract medium containing various lactic acid concentrations (0–0.8%, w/v). All data are presented as means of three replicate tubes, and standard deviations are shown as error bars. OD: optical density.
Discussion
Sake production could be divided into two major stages: the rice koji production process and the fermentation process. The rice koji production process is shown in Supplemental Figure S1 and Table S2. In the fermentation process, rice, rice koji, and water are added in sake mash on days 1, 3 and 4 after the start of sake production as shown in Supplemental Table S3. The fermentation process proceeds for approximately 20 days, and after which sake mash is pressed, filtered, in most cases pasteurized to complete the process [Citation5]. In particular, the rice koji production process has a high risk of contamination and it significantly affects the control of product quality. However, there has been limited research conducted on the microbiological aspects of this process. Therefore, the clarification of the microbiological dynamics in the rice koji production process would contribute to improving the quality of the entire sake production process.
The predominant bacteria of rice koji samples were considered as Staphylococcus spp. (). Although many researchers have reported the changes in microbial flora during the brewing process analyzed by next-generation sequencing [Citation5, Citation8, Citation11–14], those reports did not show the bacterial flora in multiple rice koji samples. However, presence of Bacillus spp. and Staphylococcus spp. was detected in those studies and seemed to be derived from rice koji in the early stage of the brewing process, suggesting microbial contamination begins in the rice koji. The present study is, to our knowledge, the first to present the results of bacterial flora analysis using multiple rice koji samples. Since the sequences obtained by next-generation sequencing are short, species-level identification is difficult. Therefore, we performed bacterial identification via a classical culture method and nearly full sequencing of the 16S rRNA gene.
As shown in Supplemental Figure S1 and Table S2, in the typical rice koji production process, the rice is treated with steam at 100 °C for approximately 50 min, which kills most of the bacteria in the rice. However, the temperature of the subsequent process is maintained at around 30 °C (), which is suitable for bacterial growth. Of the samples, the initial number of bacteria detected in the sample from brewery B was higher compared to that detected in the sample from brewery A, which could have affected the number of bacteria detected in the subsequent stages of the process. It is likely that the bacteria detected in standard assay agar originated from the air or the skin of a worker during the manual mixing. The predominant bacterial colonies were yellow and had a homogeneous morphology. The cell numbers of A. oryzae did not differ from 0 min (Tokomomi) to 2935 min, i.e. 48.9 h (Dekoji), for the koji samples from breweries A and B. This indicates that A. oryzae had not yet reached the spore-producing stage. The detected yeasts are thought to be spontaneous components, i.e. house yeast; however, since the cell numbers were very low, they posed only a low risk of abnormal fermentation in the sake mash.
Isolation of S. gallinarum from Asian foods such as broad bean paste and Japanese and Chinese fermented soybean pastes, has been reported [Citation15–17]. Additionally, S. gallinarum is known to produce a lanthionine-containing peptide antibiotic called gallidermin [Citation18, Citation19]. Unfortunately, no antimicrobial activity was detected in the strains tested in the present study (data not shown), but S. gallinarum may become the dominant strain by exhibiting faster growth than other bacteria in the rice koji production process. Koyanagi et al. reported that certain microorganisms such as S. gallinarum are predominantly detected in the early stages of sake (Yamahai-moto) fermentation, and then as fermentation progresses, S. gallinarum is no longer detected [Citation8]. Additionally, other isolated bacteria as minor populations belonged to the following genera Micrococcus, Kocuria, Bacillus, Brachybacterium, Microbacterium, Curtobacterium and Deinococcus.
During the approximately 2-days production time of rice koji, it is mixed by hand to enhance the growth of A. oryzae. This manual mixing increases the chances of bacterial contamination from the worker. It was also confirmed that all mixing of the samples used in the present study was performed without the use of plastic gloves. Notably, a particular bacterium became dominant even in samples from distinct breweries, which could be attributed to temperature selection or differences in growth speed.
S. gallinarum has been reported to be responsible for the odor associated with 4-vinylguaiacol in sake [Citation4], and excessive growth of this bacterium may result in a foul odor. Therefore, preventing contamination by S. gallinarum during rice koji production could reduce the risk of an off-odor in the resultant sake.
The phenomenon of bacterial species appearing in the early stages of fermentation and then disappearing as fermentation progresses is not uncommon. In this study, we analyzed the bacterial flora occurring during the production of rice koji, and identified the isolates by culture methods and sequencing. Staphylococcus gallinarum was found to be the dominant species during the production process, but its growth was effectively suppressed by techniques currently used by sake breweries. These results indicate that the techniques used by sake breweries in Japan are effective for controlling contamination by spoilage microorganisms [Citation4]. These bacteria might originate from the skin of the sake brewer as one possibility, because sake brewing involves human handiwork. This assumption is in agreement with several reports of Staphylococcus gallinarum [Citation15–17, Citation20, Citation21] detected in the bacterial flora analysis on fermented food.
Conclusions
We revealed the dynamics of microorganisms by amplicon sequencing and almost complete length of the V3-V4 region of 16S rRNA gene from isolates. The predominant taxon in each sample during the rice koji production process was identified as Staphylococcus gallinarum. Interestingly, the control measures in the traditional sake production process can effectively inhibit the growth of undesirable bacteria such as S. gallinarum that emerge during the production of rice koji. We anticipate that the data from this study will contribute to a better understanding of the microbial control methods used during rice koji production.
Author’s contribution
M.H. and H.Y. supervised the team and the research. K.N., M.H. and H.Y. designed the methodology and the research. K.N., T.H., K.H., N.A., M.H. and H.Y. collected the data and performed data analysis. K.N., Y.N. and S.K. performed statistical analysis. K.N., T.H., K.H. and N.A. provided the study materials, reagents, materials, laboratory samples, instrumentation, computing resources. Y.N., and S.K. provided instrumentation, computing resources. K.N., T.H., K.H., N.A., M.H. and H.Y. verified that the results are reproducible. K.N. prepared and created figures, tables and wrote the manuscript. Y.N., S.K., M.H. and H.Y. critically reviewed the manuscript. M.H. and H.Y. performed the project administration. All authors read and approved the manuscript.
Supplemental Material
Download PDF (955.4 KB)Acknowledgements
The authors thank Bioengineering Lab. Co., Ltd. (Sagamihara, Japan) for amplicon sequencing, Yukitaka Iizumi for the 16S rRNA gene sequencing data, FASMAC (Kanagawa, Japan) for direct sequence and all members of the Food & Liquor and Biotechnology Section of Yamanashi Industrial Technology Center for the opportunity to conduct this research.
Data availability statements
All data that support the findings from this study are available from the corresponding author [K.N.] upon reasonable request.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Yamashita H. Koji starter and koji world in Japan. J Fungi (Basel). 2021;7(7):1–11. doi: 10.3390/jof7070569.
- Nakagawa T, Shimada M, Mukai H, et al. Detection of alcohol-tolerant hiochi bacteria by PCR. Appl Environ Microbiol. 1994;60(2):637–640. doi: 10.1128/aem.60.2.637-640.1994.
- Akaike M, Miyagawa H, Kimura Y, et al. Chemical and bacterial components in sake and sake production process. Curr Microbiol. 2020;77(4):632–637. doi: 10.1007/s00284-019-01718-4.
- Ito T, Konno M, Shimura Y, et al. Formation of guaiacol by spoilage bacteria from vanillic acid, a product of rice koji cultivation, in Japanese sake brewing. J Agric Food Chem. 2016;64(22):4599–4605. doi: 10.1021/acs.jafc.6b01031.
- Bokulich NA, Ohta M, Lee M, et al. Indigenous bacteria and fungi drive traditional kimoto sake fermentations. Appl Environ Microbiol. 2014;80(17):5522–5529. doi: 10.1128/AEM.00663-14.
- Hui W, Hou Q, Cao C, et al. Identification of microbial profile of koji using single molecule, real-time sequencing technology. J Food Sci. 2017;82(5):1193–1199. doi: 10.1111/1750-3841.13699.
- Zhao X, Wang Y, Cai W, et al. High‑throughput sequencing‑based analysis of microbial diversity in rice wine koji from different areas. Curr Microbiol. 2020;77(5):882–889. doi: 10.1007/s00284-020-01877-9.
- Koyanagi T, Nakagawa A, Kiyohara M, et al. Tracing microbiota changes in yamahai-moto, the traditional Japanese sake starter. Biosci Biotechnol Biochem. 2016;80(2):399–406. doi: 10.1080/09168451.2015.1095067.
- Yamamura H, Harayama S. Method for selective isolation of mycobacteria using olive oil emulsified with SDS. Biosci Biotechnol Biochem. 2007;71(6):1553–1556. doi: 10.1271/bbb.60687.
- Tamura T, Hatano K. Phylogenetic analysis of the genus Actinoplanes and transfer of Actinoplanes minutisporangius Ruan et al. 1986 and ‘Actinoplanes aurantiacus’ to Cryptosporangium minutisporangium comb. nov. and Cryptosporangium aurantiacum sp. nov. Int J Syst Evol Microbiol. 2001;51(Pt 6):2119–2125. doi: 10.1099/00207713-51-6-2119.
- Hongbin W, Quanzeng W, Shuqi G, et al. Metagenomic profiling of the bacterial community changes from koji to mash stage in the brewing of soy sauce. Pol J Microbiol. 2017;66(4):537–541. doi: 10.5604/01.3001.0010.7097.
- Terasaki M, Fukuyama A, Takahashi Y, et al. Bacterial DNA detected in Japanese rice wines and the fermentation starters. Curr Microbiol. 2017;74(12):1432–1437. doi: 10.1007/s00284-017-1337-4.
- Terasaki M, Miyagawa S, Yamada M, et al. Detection of bacterial DNA during the process of sake production using Sokujo-Moto. Curr Microbiol. 2018;75(7):874–879. doi: 10.1007/s00284-018-1460-x.
- Tsuji A, Kozawa M, Tokuda K, et al. Robust domination of Lactobacillus sakei in microbiota during traditional Japanese sake starter Yamahai-Moto fermentation and the accompanying changes in metabolites. Curr Microbiol. 2018;75(11):1498–1505. doi: 10.1007/s00284-018-1551-8.
- Kim TW, Lee JH, Park MH, et al. Analysis of bacterial and fungal communities in Japanese- and Chinese-fermented soybean pastes using nested PCR–DGGE. Curr Microbiol. 2010;60(5):315–320. doi: 10.1007/s00284-009-9542-4.
- Jia Y, Niu CT, Xu X, et al. Metabolic potential of microbial community and distribution mechanism of Staphylococcus species during broad bean paste fermentation. Food Res Int. 2021;148:110533. doi: 10.1016/j.foodres.2021.110533.
- Lin H, Zhou B, Zhao J, et al. Insight into the protein degradation during the broad bean fermentation process. Food Sci Nutr. 2022;10(8):2760–2772. doi: 10.1002/fsn3.2879.
- Kellner R, Jung G, Horner T, et al. Gallidermin: a new lanthionine-containing polypeptide antibiotic. Eur J Biochem. 1988;177(1):53–59. doi: 10.1111/j.1432-1033.1988.tb14344.x.
- Götz F, Perconti S, Popella P, et al. Epidermin and gallidermin: staphylococcal lantibiotics. Int J Med Microbiol. 2014;304(1):63–71. doi: 10.1016/j.ijmm.2013.08.012.
- Dhanya Raj CT, Kandaswamy S, Suryavanshi MV, et al. Genomic and metabolic properties of Staphylococcus gallinarum FCW1 MCC4687 isolated from naturally fermented coconut water towards GRAS assessment. Gene. 2023;867:147356. doi: 10.1016/j.gene.2023.147356.
- Nemoto Y, Haraga H, Kimura S, et al. Occurrence of staphylococci in the oral cavities of healthy adults and nasal–oral trafficking of the bacteria. J Med Microbiol. 2008;57(Pt 1):95–99. doi: 10.1099/jmm.0.47561-0.
