Abstract
Tachisuzuka, a rice cultivar grown for whole-crop silage, is characterized by the small-panicle trait and high-stem sugar content. To investigate the interrelationship between the two features, we attempted to identify the gene responsible for the small-panicle trait in Tachisuzuka, and also to examine the function of the gene using a knockout mutant line. A functionally disruptive deletion of the nucleotide sequence was found in the gene SP1 (Short-Panicle 1; Os11g0235200) in Tachisuzuka, which has been reported as a candidate gene for the small-panicle trait. A gene knockout mutant of SP1 obtained from the cultivar Nipponbare showed a small-panicle phenotype similar to that observed in Tachisuzuka. However, soluble sugar content in the stem did not increase in the knockout line, whereas starch content increased significantly. Overall, disruption of SP1 is responsible for the small-panicle phenotype of Tachisuzuka, but it is only partially associated with the high-stem sugar content.
Keywords:
Classification:
Introduction
During the last decade, the Japanese government has encouraged farmers to cultivate rice for whole-crop silage (WCS), and breeding efforts have focused on developing rice cultivars having favorable traits for WCS, such as high straw yield and high-stem sugar content. Among such cultivars, Tachisuzuka was developed through introducing the small-panicle trait into an existing cultivar (Kusanohoshi) used for WCS (Matsushita et al., Citation2011; Supplementary Figure S1a). The small-panicle trait is expected to reduce the number of rice grains, which are indigestible for cattle, and simultaneously promote higher allocation of assimilates to the stems. These expectations have been successfully realized in Tachisuzuka, and more recently in Tachiayaka, another WCS cultivar bred using the same breeding strategy (Matsushita et al., Citation2014). The small-panicle trait of both WCS cultivars was donated from a common parental mutant line Gokutansui (00Kosen11). Regarding the responsive gene for the short-panicle phenotype, Matsushita et al. (Citation2011) pointed out that the panicle of Gokutansui (00Kosen11) is morphologically similar to that of the short panicle 1 (sp1) mutant, which was identified as a disruption mutant of a putative gene for a peptide transporter (Li et al., Citation2009). However, the structure and functionality of the SP1 gene have not been explored for either Gokutansui (00Kosen11) or the two descendant WCS cultivars.
In addition to the low grain/straw ratio conferred by the small panicle trait, the two WCS cultivars accumulate a high concentration of soluble sugars in their stems, which is an outstanding feature contributing to better fermentation quality of the silage (Matsushita et al., Citation2011, Citation2014). Reduction of sink size by spikelet thinning of rice has been reported to increase the contents of both soluble sugar and starch in the stems (Murata et al., Citation1997; Wada et al., Citation1993). Hence, it is reasonable to attribute the high stem sugar content of the two WCS cultivars to their small-panicle trait. However, the soluble sugar content in the stems of Tachisuzuka is extraordinarily high (Matsushita et al., Citation2011; Supplementary Figure S1b), and thus it cannot be excluded that the responsive mutation of the small-panicle trait has pleiotropic effects that promote sugar accumulation in the stem.
In this study, we examined the structure of the SP1 gene of both Tachisuzuka and Tachiayaka to confirm that disruption of SP1 causes their small-panicle trait. In addition, the effect of SP1 gene disruption on stem sugar content was examined using a knockout mutant line.
Materials and methods
Plant materials and growth conditions
Seeds of four cultivars of rice (Oryza sativa L.), Hoshiaoba, Kusanohoshi, Tachiayaka, and Tachisuzuka, were obtained from NARO Western Region Agricultural Research Center. For DNA extraction, the rice seeds were sown in plastic pots filled with nursery soil and grown in a glasshouse until the fifth leaf elongation stage. The fully elongated fourth leaf and the elongating fifth leaf were sampled and stored at −20 °C until used.
The SP1 knockout mutant of rice (cultivar Nipponbare) was obtained from the Rice Tos17-retrotransposon Insertion Mutant Panel (http://tos.nias.affrc.go.jp/; Miyao et al., Citation2003), developed at the National Institute of Agrobiological Resources (the present National Agricultural and Food Research Organization). The homozygous SP1 mutant lines, referred to as sp1, and the homozygous wild-type lines, referred to as WT, were selected from the same Tos17 insertion line, ND0393, by genomic PCR analysis. The genotyping procedures were essentially the same as those described by Okamura et al. (Citation2013), using gene-specific primers for SP1 and Tos17, the nucleotide sequences of which are listed in Supplementary Table S1. These primers were designed to produce a 274-bp product from the wild-type allele and a 198-bp product from the Tos17-inserted mutant allele.
For phenotypic analyses, the sp1 and WT plants were grown in an experimental paddy field at the Institute for Sustainable Agro-ecosystem Services (ISAS), The University of Tokyo (35°44′N, 139°32′E, altitude: 58 m), Tokyo, Japan, during the summer months in 2013. One-month-old seedlings, grown in a greenhouse, were transplanted into the paddy field in late May. The plant density was 22.2 hills m−2 (hill spacing: 0.30 m × 0.15 m) with one seedling per hill, and the plot size was 1.5 m × 2.1 m. Compound fertilizer for paddy fields was applied at 6 g N m−2, 8 g P2O5 m−2, and 9 g K2O m−2 as a basal dressing. During harvest (6 weeks after heading), the leaf sheaths and internodes of main stems were sampled from four individual hills; the leaf sheath of the leaf just below the flag-leaf (LS2), the leaf sheath of the second leaf below the flag-leaf (LS3), the second-top internode (Int2), and the third-top internode (Int3). These stem parts were frozen immediately in liquid nitrogen, and stored at −80 °C until used. Panicles attached to the same main stems were also sampled and weighed after drying in air for 1 week.
DNA extraction and analysis of SP1 gene structure
Genomic DNA was extracted from young leaves using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany) following the manufacturer’s instruction. After quantification by fluorometric assay (Qubit dsDNA HS Assay Kit; Thermo Fisher Scientific, Waltham, MA, USA), 5 ng of the DNA sample was used as a template for each PCR amplification using a high-fidelity DNA polymerase (PrimeScript GXL; TAKARA BIO Inc., Shiga, Japan) with a primer pair, OsSP1-L1 and R1. The nucleotide sequence of the amplified DNA fragment was determined using an automatic DNA sequencer (3730xl; Applied Biosystems, Foster City, CA, USA) and assembled using computer software (GENETYX ver10; Genetyx Co., Tokyo, Japan) to analyze the gene structure. The nucleotide sequences of the PCR primers used are listed in Supplementary Table S1.
Determination of carbohydrate content
Frozen leaf-sheath and internode samples were milled under cryogenic conditions using a Multi-Beads Shocker (Yasui Kikai, Osaka, Japan). A portion (~100 mg) of the resultant tissue powder was used for enzymatic quantification of starch, sucrose, glucose, and fructose, according to the methods described by Okamura et al. (Citation2013).
Results and discussion
Structure of the SP1 gene in Tachisuzuka
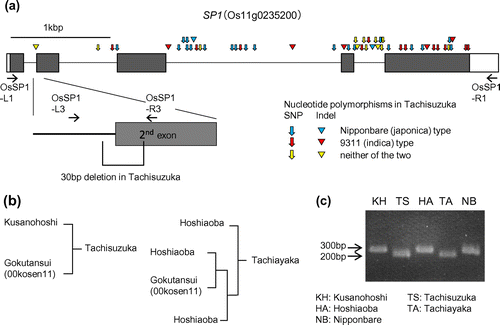
As the first step to determine whether the small-panicle phenotype of Tachisuzuka is conferred by mutation of SP1, the nucleotide sequence of SP1 in Tachisuzuka was examined and compared with corresponding sequences in the genome databases of japonica (cultivar Nipponbare; RAP_DB, http://rapdb.dna.affrc.go.jp) and indica rice (germplasm 9311; RIS, http://rice.genomics.org.cn/rice). In the genome databases, SP1 corresponds to a gene comprising five exons on chromosome 11, with locus ID Os11g0235200 (RAP_DB) or BGIOSGA034245 (RIS), which is annotated as a putative gene encoding a peptide transporter. Comparison of the SP1 sequences from japonica and indica databases revealed that there are 42 SNPs and 7 indels in the open-reading frame between the two rice subspecies (Figure a). In the SP1 sequence of Tachisuzuka, some of these SNPs and indels are japonica-type alleles, whereas others are indica-type alleles. In addition to these SNPs and indels, the SP1 gene of Tachisuzuka contains 12 nucleotide polymorphisms that are present in neither of the two reference sequences and, importantly, one of these polymorphisms is characterized by a 30-nucleotide deletion that straddles the exon/intron junction of the 5′-end of the second exon, which presumably results in dysfunction of the gene (Figure (a)). This finding suggests that disruption of SP1 is responsible for the small-panicle trait of Tachisuzuka. To further confirm this, we examined the SP1 gene structure of Tachiayaka, another small-panicle WCS cultivar, having Gokutansui (00Kosen11) as a great-grandfather (Figure (b)). In addition, the seed-parents of the two WCS cultivars, Kusanohoshi and Hoshiaoba, were also examined. In these cultivars, the 30-nucleotide deletion was detected by the length of PCR product amplified by the specific primers spanning the deletion (Figure (c)). Compared to their seed-parents, shorter DNA fragments with seemingly identical gel mobility were amplified in Tachiayaka and Tachisuzuka, suggesting that a common mutated SP1 derived from Gokutansui (00Kosen11) has been inherited in the two WSC cultivars. Collectively, it can reasonably be concluded that the small-panicle trait of Tachisuzuka and Tachiayaka is conferred by disruption of the SP1 gene inherited from the pollen parent.
Figure 1. Analysis of the SP1 gene structure in Tachisuzuka and related cultivars. (a) Exon/intron structure of SP1 (Os11g0235200). Boxes indicate exons, and the coding region is shown by the shadow. The nucleotide deletion in Tachisuzuka on the exon/intron boundary is magnified. The positions and the types of the nucleotide polymorphisms in Tachisuzuka are show by arrows (SNPs) and triangles (indels) in three different colors to indicate the types of alleles. The positions of the PCR primers used are also shown (OsSP1-L1, L3, R1, and R3; see Supplementary Table S1 for detail). (b) Genealogy of Tachisuzuka and Tachiayaka (modified from Matsushita et al. Citation2011, Citation2014). (c) PCR analysis to examine the presence of the nucleotide deletion in the SP1 gene of Tachisuzuka, Tachiayaka, and their relatives, with Nipponbare as a control. The primers OsSP1-L3 and R3 that span the deletion were used.
Disruption of the SP1 gene and stem soluble sugar content
From the rice Tos17-retrotransposon insertion mutant line ND0393, we isolated homozygous SP1 mutant lines (sp1), in which Tos17 is inserted into the second exon of the SP1 gene (Figure (a) and (b). The panicles of sp1 were shorter than those of the WT, being very similar to those in Tachisuzuka and Kusanohoshi (Figure (c), Supplementary Figure S1a). The panicle weight of sp1 was less than half that of the WT (Figure (d)). These observations confirm that disruption of the SP1 gene results in the small-panicle phenotype.
Figure 2. The SP1-KO mutant of Nipponbare used in this study. (a) Gene structure of SP1 and the position of the Tos17 insertion in the ND0393-derived knockout mutant. The names and positions of the primers used for genotyping are indicated with arrows (see Materials and Methods for details). (b) Representative result of genomic PCR analysis for genotyping to select sp1 and WT rice plants. (c) The panicles of sp1 and WT at harvest. (d) Comparison of the panicle weight of sp1 and WT at harvest. Data represent the means and standard errors. Note that the difference between the two lines was significant, as determined by Student’s t-test (p < 0.001, n = 5).
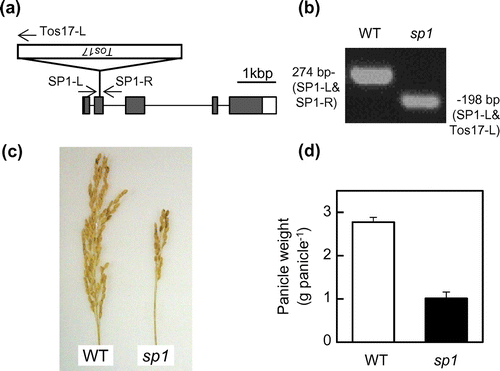
Table summarizes the concentrations of starch and soluble sugars (sucrose, glucose, and fructose) in four different parts of the stem tissues of sp1 and WT plants at harvest. Starch concentrations in these stem tissues tended to be higher in sp1 than in the WT: the differences between the two lines were statistically significant in LS3 (leaf sheath of the second leaf below the flag-leaf) and Int3 (third-top internode), but not in LS2 (leaf sheath of the leaf just below the flag-leaf) and Int2 (second-top internode) due to variation in data among four replicates. The elevated starch levels in sp1 tissues can be explained by the hypothesis that, in sp1 plants, a considerable reduction in carbon demand by panicles promotes the production of excess carbon resources, resulting in an increase in starch reserves in the stem. In contrast, sucrose concentrations in sp1 stems were similar to or lower than those in WT stems. Although glucose and fructose concentrations were significantly higher in sp1 than in WT in all the stem tissues measured, the concentrations of these hexoses were at negligible levels in comparison with starch and sucrose concentrations. Thus, soluble sugar levels, unlike starch levels, did not increase when the sink size of panicles was reduced by the disruption of SP1. Kono (Citation2011) reported that soluble sugar concentration in the stem of Kusanohoshi was increased by panicle-thinning treatment, but that the elevated concentration was still lower than the concentration in the stem of Tachisuzuka. These insights suggest that the influence of panicle size reduction on stem soluble sugar reserves is limited in rice plants, and that extremely high-level accumulation of soluble sugars in the stem of Tachisuzuka is archived by a factor(s) other than the small panicle size.
Table 1. Comparison of starch, sucrose, glucose, and fructose concentrations in the stem tissues of the WT and SP1-KO mutant of Nipponbare.
Conclusion
Although mutation of the SP1 gene was confirmed to be responsible for the small-panicle phenotype of Tachisuzuka and Tachiayaka, the physiological function of the gene remains unresolved. However, at least in Nipponbare, disruption of SP1 does not result in high-soluble sugar content in the rice stem, although the panicle becomes smaller and stem starch content is increased. It seems reasonable that ‘demand’ can affect the amount of ‘stock,’ but not necessarily the portfolio of it. Comparative physiological studies with greater focus on the sugar metabolism of Tachisuzuka are necessary.
Supplemental data
Supplemental data for this article can be accessed here. [http://dx.doi.org/10.1080/1343943X.2016.1260484].
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was supported in part by JSPS KAKENHI [grant number 26292009] to NA and TH.
pps2016_102sr-File004.pdf
Download PDF (144.9 KB)Acknowledgments
We thank the technical support staff of the ISAS at the University of Tokyo for their assistance in the cultivation and management of rice, and Kiiko Takatsuto, NARO, for expert technical assistance. M. Okamura and Y. Hashida received a fellowship from the Japan Society for the Promotion of Science (JSPS).
References
- Kono, S. (2011). Similarity of artificial deflorated rice (common breed) to very short ear breed “Tachisuzuka” on the chemical composition. Japanese Journal of Grassland Science, 57, Extra Issue, 105**.
- Li, S., Qian, Q., Fu, Z., Zeng, D., Meng, X., Kyozuka, J., … Wang, Y. (2009). Short panicle1 encodes a putative PTR family transporter and determines rice panicle size. The Plant Journal, 58, 592–605.10.1111/tpj.2009.58.issue-4
- Matsushita, K., Iida, S., Ideta, O., Sunohara, Y., Maeda, H., Tamura, Y., … Takakuwa, M. (2011). ‘Tachisuzuka’, a new rice cultivar with high straw yield and high sugar content for whole-crop silage use. Breeding Science, 61, 86–92.10.1270/jsbbs.61.86
- Matsushita, K., Ishii, T., Ideta, O., Iida, S., Sunohara, Y., Maeda, H., & Watanabe, H. (2014). Yield and lodging resistance of ‘Tachiayaka’, a novel rice cultivar with short panicles for whole-crop silage. Plant Production Science, 17, 202–206.10.1626/pps.17.202
- Miyao, A., Tanaka, K., Murata, K., Sawaki, H., Takeda, S., Abe, Y., … Hirochika, H. (2003). Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. The Plant Cell, 15, 1771–1780.10.1105/tpc.012559
- Murata, T., Hosaka, Y., Hirano, M., & Kuroda, E. (1997). Carbohydrate metabolism in leaf blades and stems of rice cultivar Akitakomachi at the ripening stage. Japanese Journal of Crop Science, 66, 221–228*.10.1626/jcs.66.221
- Okamura, M., Hirose, T., Hashida, Y., Yamagishi, T., Ohsugi, R., & Aoki, N. (2013). Starch reduction in rice stems due to a lack of OsAGPL1 or OsAPL3 decreases grain yield under low irradiance during ripening and modifies plant architecture. Functional Plant Biology, 40, 1137–1146.
- Wada, Y., Miura, K., & Watanabe, K. (1993). Effects of source-sink ratio on production and senescence of rice flag leaves during the ripening period. Japanese Journal of Crop Science, 62, 547–553*.10.1626/jcs.62.547