Abstract
Recent progress in rice genomics has promoted the identification of quantitative trait loci (QTLs) associated with yield and its related traits. SPIKE, a QTL controlling spikelet number per panicle, and GPS, a QTL controlling leaf photosynthesis rate, were identical to NAL1. To assess the effect of SPIKE/GPS on yield potential, we compared DNA sequences of these alleles and conducted yield experiments in the field of Japan using the near-isogenic lines NIL-SPIKE (allele from Daringan in IR 64 genetic background), NIL-GPS (allele from Koshihikari in Takanari genetic background), and a chromosome segment substitution line, SL2115 (allele from Koshihikari in IR 64 genetic background). Despite the two SNPs in the promoter regions between Koshihikari and Daringan, both alleles were effective to increase the number of spikelets per panicle both in Takanari and IR 64 backgrounds. However, the extent of the increase was smaller and unstable in Takanari background than that in IR 64 background. In addition, SPIKE/GPS improved percentage of filled spikelets only in the IR 64 background. These results suggest that the effects of SPIKE/GPS alleles are similar but are affected by the difference of the genetic backgrounds. Because the increasing effect of spikelets number per panicle was canceled by the decrease of the number of panicles, which seems to be affected by environmental factors, none of NIL-SPIKE, SL2115, or NIL-GPS significantly out-yielded their parental cultivars. These results indicate the importance to consider genetic backgrounds and QTL-environment interaction toward the future use of SPIKE/GPS.
Keywords:
Classification:
Introduction
Plant breeding is a continuous process of crossing and selection to produce new cultivars with desirable characteristics. Rice (Oryza sativa L.), as a staple food, has long history of breeding in Asia, which has resulted in a wide range of cultivars and wide genetic diversity. One of the most crucial events in rice breeding was the dramatic increase in grain yield by the use of the semi-dwarf1 (sd1) gene in the ‘Green Revolution’ of the 1960s and 1970s (Khush, Citation2001; Peng et al., Citation1999). When the average yield of unhulled grain in tropical Asia was 1.4 t ha−1, IR 8, the first high-yielding indica cultivar (developed by IRRI in the Philippines) in tropics, produced nearly 10 t ha−1 through a high N response and high lodging resistance conferred by sd1 (De Datta et al., Citation1968). However, continuing global population growth will require a further 60–70% increase in rice production in the limited area of arable land by 2050 (Tester & Langridge, Citation2010). This increase will require improvement of rice’s yield potential through the accumulation of favorable alleles of valuable genes (The 3000 rice genomes project, Citation2014).
Rice yield is generally determined by the balance among sink size, source strength, and carbohydrate translocation. In temperate Japan, breeders developed a high-yielding indica cultivar, Takanari, in 1990 (Imbe et al., Citation2004). Takanari has a large sink size due to its large panicles, and high biomass production due in part to its high leaf photosynthetic capacity (Takai et al., Citation2006); as a result, it attained the highest yield on record, namely 11.7 t of brown rice ha−1 in Japan (Takai et al., Citation2014). In the tropics, breeders at International Rice Research Institute (IRRI) in the Philippines developed a number of high-yielding indica cultivars after the release of IR 8. One of them, IR 64, was developed in 1985 and is now widely grown in South and Southeast Asia owing to its wide regional adaptability (Uga et al., Citation2011). IR 64 produces a large number of small panicles with large grains (Peng et al., Citation2000). Breeders are continuing efforts to develop new rice cultivars with higher yielding potential than Takanari and IR 64 to ensure future food security.
Progress in rice genomics over the last two decades has enabled breeders to do molecular breeding using DNA markers and mapping information for quantitative trait loci (QTLs). With those tools, Fujita et al. (Citation2013) recently identified SPIKE, as a QTL that controls spikelet number per panicle. They confirmed that a near-isogenic line (NIL) with the SPIKE allele from tropical japonica landrace, Daringan in the IR 64 genetic background produced more number of spikelets per panicle and higher grain yield than IR 64 in the field of the IRRI under tropical condition. The results highlight the value of this allele to increase yield potential in indica cultivars. Takai et al. (Citation2013) similarly identified GREEN FOR PHOTOSYNTHESIS (GPS), which is a QTL responsible for high leaf photosynthesis in Takanari. NIL-GPS, with the GPS allele from temperate japonica cultivar, Koshihikari, in the Takanari genetic background, showed low photosynthesis rate and did not out-yield Takanari in the field of NARO Institute of Crop Science in Japan under temperate condition. Interestingly, the causal genes for both GPS and SPIKE were identical to NALLOW LEAF1 (NAL1) (Qi et al., Citation2008). Why did the same gene result in different final yields between SPIKE and GPS; yield increase in SPIKE but not increase in GPS? Possible reasons include allelic differences between Koshihikari and Daringan, different genetic backgrounds between the two indica cultivars, or different environments between the trials.
In this study, we compared sequences of NAL1 among the four rice cultivars used in the studies by Takai et al. (Citation2013) and Fujita et al. (Citation2013) to identify allelic differences. Then we conducted yield trials using Takanari, IR 64, and NILs for GPS and SPIKE to evaluate their yield potential in the same experimental paddy field in Japan. We discuss the effect of SPIKE/GPS on yield and its components toward the future use of SPIKE/GPS.
Materials and methods
Plant materials and sequence analysis
Takanari is a high-yielding indica cultivar released in Japan. Koshihikari is a leading temperate japonica cultivar in Japan with good eating quality and the largest area of cultivation during the last 30 years (Takeuchi et al., Citation2008). IR 64 is a high-yielding indica cultivar released by IRRI in the Philippines. Daringan is a tropical japonica landrace grown in Indonesia (Fujita et al., Citation2013). We amplified ~1.5-kb of the promoter region of NAL1 of each cultivar by PCR with two primer sets: 5′-CAAGAATTGGCTCCATCTCC-3′ and 5′-GCGCTGTTCTCTGTGTGTGT-3′; and 5′-CAGTGGAGATTCCCGTATCCATTTCC-3′ and 5′-GGCAACAGTTACAGCGACCTCAAGAA-3′. We amplified the cDNA region with the primer sets used by Takai et al. (Citation2013). We used 35 cycles of 1 min template DNA denaturing step at 94 °C, 1 min primer annealing step at 55 °C, and 2 min primer extension step at 72 °C in PCR program. The PCR products were sequenced with a BigDye Terminator v. 3.1 Cycle Sequencing Kit (Applied Biosystems) in an automated fluorescent laser sequencer (3130xl; Applied Biosystems).
Field experiments
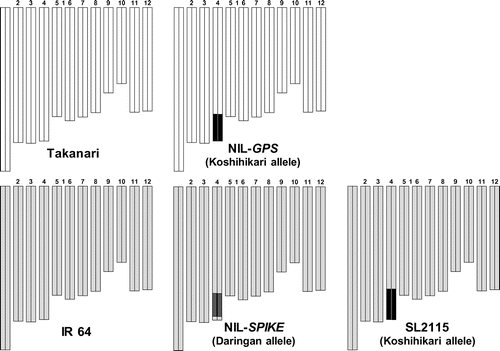
Field experiments were conducted in the experimental paddy field at the NARO Institute of Crop Science, Tsukubamirai (36°02′N, 140°04′E), Ibaraki, Japan, in 2014 and 2015. The soil is a Gleyic Fluvisol. Takanari, NIL-GPS, IR 64, and NIL-SPIKE were grown under continuous flooded conditions. NIL-GPS has a Koshihikari-derived chromosome segment that includes GPS on the long arm of chromosome 4 in the Takanari genetic background (Figure ). NIL-SPIKE has a Daringan-derived chromosome segment on the long arm of chromosome 4 in the IR 64 genetic background (Figure ). In addition to the two NILs, a chromosome segment substitution line, SL2115, which carries a Koshihikari-derived chromosome segment on the long arm of chromosome 4 in the IR 64 background (Figure ), was also grown in 2015 to compare the difference between SPIKE and GPS in the same IR 64 background.
Figure 1. Graphical genotypes of Takanari, NIL-GPS, IR 64, NIL-SPIKE, and SL2115. White bars, Takanari; black bars, Koshihikari; light gray bars, IR 64; dark gray bars, Daringan.
Seeds were sown in a seedling nursery box on 23 April 2014 and 22 April 2015, and seedlings were transplanted at one per hill on 15 May 2014 and 14 May 2015, at 22.2 hills m−2, with 15 cm between hills and 30 cm between rows. The experimental plots (18 m2 each) were arranged in a randomized complete block design with three replicates. In 2014, basal fertilizer was applied 3 days before transplanting at 12 g N m−2 as controlled-release fertilizer (4 g LP40 and 8 g LP100), 5.2 g P m−2, and 7.5 g K m−2. LP40 and LP100 release 80% of their total N content at a uniform rate by 40 and 100 days, respectively, after application at 20–30 °C. At the panicle initiation stage, 6 g N m−2 was top-dressed as LP40. In 2015, we reduced the application to 4 g N m−2 (1.3 g LP40 and 2.7 g LP100) as a basal dressing and 2 g N m−2 (LP40) as a top-dressing to evaluate the influence of N fertility to the effect of SPIKE/GPS. The applications of P and K in 2015 were the same as those in 2014.
Evaluation of biomass production, yield, and yield components
Twelve hills were sampled from each plot at the panicle initiation stage (2014), heading stage (2014 and 2015), and maturity (2014 and 2015). Heading date was defined as the date when about half of the panicles in each plot had emerged. Days-to-heading was determined as the number of days from sowing to heading. In 2014, two representative hills with an average number of tillers were selected from the 12 hills, and the leaf blades were collected to measure leaf area with an area meter (AAM-9, Hayashi Denko, Tokyo, Japan). In both years, the entire shoots were dried for 72 h at 80 °C and weighed.
Leaf chlorophyll content was measured as the SPAD value with a chlorophyll meter (SPAD-502, Konica-Minolta, Japan) at the panicle initiation stage (2014), at heading (2014 and 2015), and at maturity (2014 and 2015). The topmost fully expanded leaves from the main stem were used for measurements. Six and 12 leaves in one experimental plot were used for the measurements in 2014 and in 2015, respectively.
At maturity, plants covering 2.7 m2 (60 hills) were harvested from each plot for determination of yield and its components according to the method of Takai et al. (Citation2014). Panicles were counted and then threshed to obtain unhulled grains, which were weighed and divided equally into subsamples A and B. Approximately 40 g of unhulled grains (subsample C) was selected from subsample A and counted in an electronic seed counter (KC-10S, Fujiwara Scientific Co. Ltd., Tokyo, Japan). Spikelet number m−2 was calculated by multiplying the grain number per unit weight in subsample C by the total weight of the unhulled grains m−2. Spikelet number per panicle was calculated as the spikelet number m−2 divided by panicle number m−2. The hulls from subsample B were subsequently removed with a rice huller (25 M, Ohya Tanzo G.K. Co., Aichi, Japan), and the hulled grains were weighed to determine brown rice yield. The grains were then screened with a grain sorter (TWS, Satake Co. Ltd., Tokyo, Japan; 1.6-mm sieve size), and the single-grain weight was calculated. The filled grain percentage was calculated as the number of screened hulled grains m−2 divided by the spikelet number m−2. Brown rice yield and single-grain weight were adjusted to 15% moisture content. Sink capacity was defined as single grain weight multiplied by the number of spikelets m−2.
Statistical analysis
Statistical analyses were performed using a general linear model in the SPSS 22.0 software (IBM, Chicago, IL, USA) in each genetic background. Cultivar was considered as a fixed effect, and replication was considered as a random effect. Analysis of variance (ANOVA) was conducted to examine the effects of GPS or SPIKE on each trait in each year, because SL2115 was grown only in 2015. Significant GPS or SPIKE effects (p < .05) were tested using Student’s t-test except for IR 64 genetic background in 2015. The effects in IR 64 genetic background in 2015 was tested using Tukey’s test. Difference in each trait in each cultivar across the two years was also tested by ANOVA.
Results
Sequence comparisons among the four rice cultivars
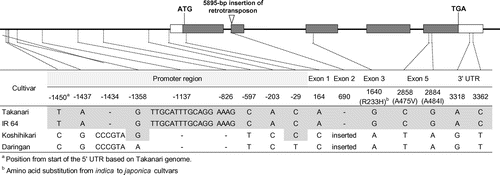
Comparison of NAL1 cDNA sequences between Takanari and Koshihikari represented six single nucleotide polymorphisms (SNPs), three of which caused amino acid substitutions, between the cultivars as well as one retrotransposon insertion in Koshihikari (Figure ). The sequences of IR 64 and Daringan were identical to those of Takanari and Koshihikari, respectively (Figure ). In the promoter region, there were nine polymorphisms among the four cultivars. The sequences of Takanari and IR 64 were identical (Figure ). Koshihikari and Daringan had seven and nine polymorphisms, respectively, against the two indica cultivars, which indicate that two SNPs were existed between Koshihikari and Daringan in the promoter region (Figure ). The base sequences of the two SNPs in Koshihikari were the same as those in the two indica cultivars.
Climate conditions and growth durations
Mean temperatures during the growth period were similar in 2014 and 2015 (Figure (A)). They showed a gradual increase until heading in mid-August, and then a gradual decrease during grain filling until maturity. Solar radiation also showed similar patterns in 2014 and 2015, except for a remarkable decline in early July 2015 (Figure (B)). Radiation levels were high after transplanting in May and during the reproductive period between mid-July and mid-August. Takanari and NIL-GPS headed at ~105 days after sowing, and IR 64, NIL-SPIKE, and SL2115 headed at 108–114 days after sowing (Table ). Accordingly, total growth durations were longer in IR 64, NIL-SPIKE, and SL2115 than in Takanari and NIL-GPS (Table ).
Figure 3. (A) Mean temperature and (B) solar radiation during the rice growth period measured in the experimental field.
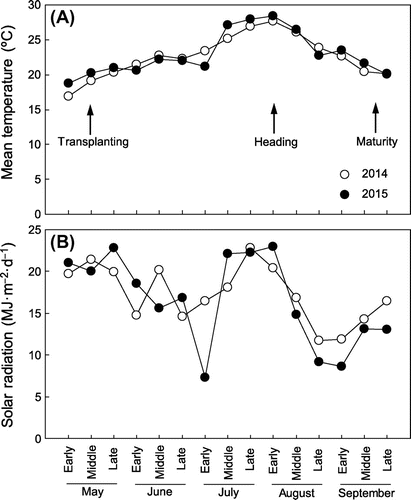
Table 1. Days-to-heading and growth duration in five cultivars in 2014 and 2015.
Comparisons of yield and its components
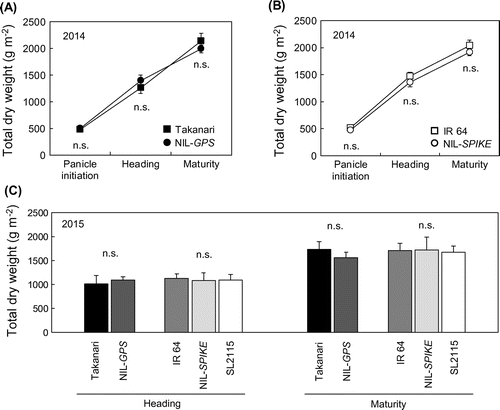
In 2014, Takanari yielded 1241 g m−2 of unhulled grain and 919 g m−2 of brown rice (Table ). The yield of IR 64 was lower than that in Takanari, namely 781 g m−2 of unhulled grain and 562 g m−2 of brown rice. In 2015, yields of unhulled grain and brown rice in Takanari were 13 and 10% lower than those in 2014 due to low N application, respectively, while the yield in IR 64 did not decrease compared with that in 2014. The yields in NIL-GPS and NIL-SPIKE also showed the similar pattern to those in their parental cultivars in both years. Consequently, NIL-GPS, NIL-SPIKE, and SL2115 did not out yield their parental cultivars in both years (Table ).
Table 2. Yield and yield components in five cultivars in 2014 and 2015.
For yield components, our field experiments confirmed that SPIKE controls the number of spikelets per panicle (Fujita et al., Citation2013): the number was significantly larger in NIL-SPIKE than in IR 64 in 2014 (Table ). NIL-GPS and SL2115 also produced a significantly greater number of spikelets per panicle than their parental cultivars in 2015. The extent of the increase was smaller in Takanari background than that in IR 64 background. On the other hand, there was no difference in the number of spikelets m−2 among them except for between Takanari and NIL-GPS in 2014, because NILs and SL2115 had fewer panicles than their parental cultivars. Takanari and NIL-GPS, with the Takanari genetic background, produced more spikelets m−2 than IR 64, NIL-SPIKE, and SL2115, with the IR 64 genetic back ground, because of their large panicles (~200 vs. ~110 spikelets per panicle). However, decreased rate of spikelet number m−2 in Takanari and NIL-GPS in 2015 relative to in 2014 due to low N application was >20%, while the decreased rate in IR 64 and NIL-SPIKE was <20%.
We found significant differences in single-grain weight and percentage of filled spikelets between the parental cultivars and the NILs or SL2115 (Table ). The single-grain weight of brown rice in both NILs and SL2115 was significantly lower than that in their parental cultivars except for between Takanari and NIL-GPS in 2015. NIL-SPIKE showed relatively higher percentage of filled spikelets than IR 64, while NIL-GPS did not show higher percentage of filled spikelets than Takanari.
The comparison between NIL-SPIKE and SL2115 is useful to examine the difference of the effects between SPIKE and GPS in the same IR 64 genetic background. No significant difference was observed for yield and its components except for single-grain weight between NIL-SPIKE and SL2115 in 2015, indicating that the effects on yield and its component are similar between SPIKE and GPS.
Eventually, NIL-GPS, NIL-SPIKE, and SL2115 did not have a significantly larger sink capacity than their parental cultivars (Table ). The number of spikelets m−2 was significantly correlated with brown rice yield (r = .77, p < .05; Table ).
Table 3. Correlation coefficient (Pearson’s r) between brown rice yield and the three yield components.
Comparisons of leaf area index (LAI), chlorophyll content, and biomass production
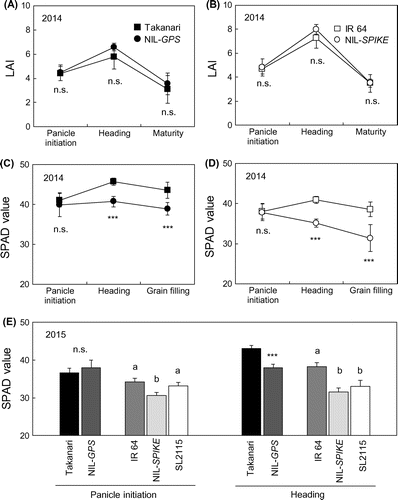
GPS controls leaf size and chlorophyll content (Takai et al., Citation2013). LAI peaked at heading in all four cultivars in 2014 (Figure (A) and (B)). At heading, IR 64 had 25% higher LAI than Takanari, and NIL-SPIKE and NIL-GPS had 11 and 14% higher LAI than IR 64 and Takanari, respectively, although the differences were not significant. The SPAD value, an index of leaf chlorophyll content, differed significantly between the parental cultivars and the NILs or SL after heading: Takanari had a higher SPAD value than IR 64, and the NILs and SL2115 had a significantly lower SPAD value than the parental cultivars at heading (Figure (C)–(E)). The differences remained significant during grain filling (Figure (C) and (D)).
Figure 4. Change of (A, B) leaf area index (LAI) in 2014 and (C–E) chlorophyll content (as the SPAD value) in the topmost fully expanded leaves in (C, D) 2014 and (E) 2015 in five cultivars. *** shows p < .001 by student’s t test between the parent and its NIL in 2014 and in the Takanari background in 2015. Different letter shows p < .05 by Tukey’s test among IR 64, NIL-SPIKE, and SL2115 in 2015. n.s. shows not significant.
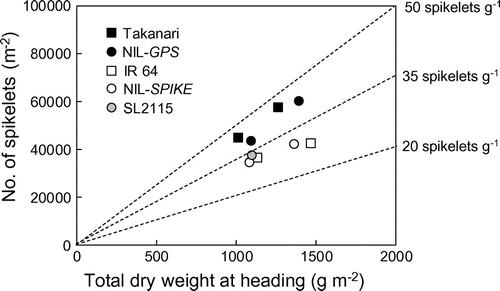
There was no difference in biomass production between Takanari and IR 64 or between parental cultivars and the NILs or SL2115 in either year (Figure (A)–(C)). Despite this, Takanari and NIL-GPS produced 39–45 spikelets per unit biomass, but IR 64, NIL-SPIKE, and SL2115 produced 29–34 (Figure ).
Discussion
The effect of sequence differences within alleles of NAL1
In this study, we found no DNA sequence difference in the promoter and cDNA regions between Takanari and IR 64 but found some differences between Takanari and Koshihikari as well as IR 64 and Daringan (Figure ). Within these differences, one of the most remarkable differences is a retrotransposon insertion in the second exon of NAL1 both in Koshihikari and Daringan (Figure ). Although an insertion of retrotransposon generally destroys the inserted gene and leads to loss-of-function of the gene, NAL1 was expressed in both Koshihikari and Daringan in previous studies and seems to be functional alleles (Fujita et al., Citation2013; Takai et al., Citation2013). Expression of many splice variants in NAL1 transcripts observed in Koshihikari and Takanari is another conspicuous point of this gene (Takai et al., Citation2013). However, it is still not clear how the retrotransposon insertion and splice variants affected the gene expression level and further expression analysis might be necessary.
Another candidate of sequence difference that may influence the effect of NAL1 is the SNPs in the cDNA region. Recently, Taguchi-Shiobara et al. (Citation2015) suggested that an R233H amino acid substitution in the third exon may explain the phenotypic difference for leaf morphology between indica and japonica cultivars. The indica cultivars Takanari and IR 64 carry the R-233 allele, and the japonica cultivars Koshihikari and Daringan carry the H-233 allele. This suggests that the amino acid substitution may cause the phenotypic difference both in Daringan SPIKE substitution in the IR 64 background and Koshihikari GPS substitution in the Takanari background.
The factor that may cause the difference between Koshihikari and Daringan is the two SNPs in the promoter region in NAL1 (Figure ). It is suggested that either or both SNPs may be related to the higher gene expression level of the Daringan allele than the IR 64 allele in young panicles (Fujita et al., Citation2013) although no increase of NAL1 expression was observed in young flag leaves between Koshihikari and Takanari (Takai et al., Citation2013). However, these two experiments were conducted at the different organs and stages. Comparisons of gene expression level among the parental cultivars or NILs in the same stage organ such as young panicles are necessary to conclude whether the two SNPs are the key factors to regulate the gene expression or not.
Effect of SPIKE/GPS on yield and its components
In yield trials, Takanari produced larger panicles (a larger number of spikelets per panicle) than IR 64, which resulted in a larger number of spikelets m−2 and thus higher yield than IR 64 across the two years (Table ). However, Takanari remarkably declined yield in 2015 compared to 2014, while yield level in IR 64 was consistently stable across the two years. Main difference between the two years is the N application level; 18 g N m−2 in 2014 and 6 g N m−2 in 2015. Takanari seems to require more N, but IR 64 seems to be stable under various environmental conditions including poorly fertilized one.
Our results suggest that the effects of SPIKE/GPS may be influenced by the difference in genetic backgrounds. Both Koshihikari GPS and Daringan SPIKE alleles affected the increase of the number of spikelets per panicle and decrease of chlorophyll content of leaves irrespective of the genetic backgrounds as well as regardless of the two SNPs. However, the effect of increasing the number of spikelets per panicle was smaller in the Takanari genetic background than in the IR 64 genetic background. The alleles of SPIKE/GPS might not be effective in the genetic background of high yielding indica cultivars with low panicle number and big panicles such as Takanari because Takanari already attains large panicles at least by the alleles of GN1a and APO1 for the direction of increasing spikelet number (Takai et al., Citation2014) and hence already has higher sink capacity than IR 64. Therefore, there may not be an enough room to raise the number of spikelets in the Takanari background by SPIKE/GPS.
We also observed that SPIKE/GPS in the IR 64 genetic background improved the percentage of filled spikelets in addition to the number of spikelets per panicle. On the contrary, no increase of the percentage of filled spikelets was observed in the Takanari background. Lack of surplus supply of carbohydrate caused by the high sink capacity in Takanari might be one of the reasons of the low improvement of filled spikelet percentages. These results also confirm that the effects of SPIKE/GPS on the yield components are affected by the difference in genetic backgrounds. SPIKE/GPS may be more useful to improve the yield components in indica cultivars with small panicles such as IR 64 rather than those with large panicles such as Takanari.
Despite the effects of SPIKE/GPS to increase the number of spikelets per panicle, none of NIL-SPIKE, SL2115, or NIL-GPS significantly out-yielded their parental cultivars under the present field conditions regardless of the amount of N applications (Table ). No increase of final yield is considered to be caused by a trade-off between the number of spikelets per panicle and the number of panicles, which resulted in the similar number of spikelets m−2 and the similar sink capacity, which significantly correlated to the final yield (Table ). This indicates why the NILs and SL2115 did not out-yield their parental cultivars in this study.
The reason why the effect of SPIKE differs in tropics and temperate Japan
So why did NIL-SPIKE out-yield IR 64 in the study by Fujita et al. (Citation2013)? A possible reason is the difference in vegetative growth period between their study and ours. Days-to-heading in IR 64 and NIL-SPIKE was approximately 80–90 days in the Philippines (see Figure S2 of Fujita et al., Citation2013) and 108–114 days in this study (Table ). Under the tropical climate conditions such as short day and high temperature, acceleration of flowering restricts the period of vegetative growth. In such condition, IR 64 did not produce a greater number of panicles per plant compared with NIL-SPIKE (see Figure S2 of Fujita et al., Citation2013). On the other hand, the difference of producing tillers and hence panicles between IR 64 and NIL-SPIKE will become bigger under prolonged vegetative growth condition such as in this study, and therefore the effects of SPIKE to increase the number of spikelets per panicle was canceled by the lower number of panicles (Table ). These results suggest that SPIKE may have an advantage in increasing the number of spikelets m−2 under the conditions that tillering is restricted due to such as short vegetative growth period as observed in Fujita et al. (Citation2013). If this principle is true, SPIKE may be also useful to the paddy field where vegetative growth period is restricted due to late transplanting after the harvesting of winter crops such as wheat or barley. Also, under the conditions that plants cannot attain enough tillering, growing NIL-SPIKE with high plant density may compensate for the fewer panicles m−2 and may have high sink capacity. Further trials to apply SPIKE/GPS to increase grain yield are necessary under conditions where plants can have adequate vegetative growth period such as in Japan.
Another possible reason is that SPIKE/GPS is effective under low yield level conditions as observed in Fujita et al. (Citation2013). Yield level of IR 64 in Fujita et al. (Citation2013) was lower than that in our low fertility experiment in 2015. In such low soil fertility condition in Fujita et al. (Citation2013), tillering and thus the number of panicle formation are also restricted. Consequently, the effect of increasing spikelet number per panicle by SPIKE/GPS may have advantage under such low yield level conditions by low soil fertility.
Effect of SPIKE/GPS on biomass production
Large biomass production is also an important trait for achieving high yield potential (Dingkuhn et al., Citation2015; Peng et al., Citation2008; Takai et al., Citation2009). Although the NILs had a lower leaf chlorophyll content and a relatively greater LAI than the parental cultivars at heading (Figure ), biomass production was not affected by GPS or SPIKE (Figure ). This absence of an effect of GPS or SPIKE on biomass production may be another reason why the NILs and SL2115 did not out-yield the parental cultivars. Although there is no information on biomass production in the experiments by Fujita et al. (Citation2013), NIL-SPIKE is possible to increase biomass production under tropical conditions. As described above, both IR 64 and NIL-SPIKE produced the similar panicle number in the Philippines. Under such conditions, the effect of SPIKE to enlarge each leaf in each tiller may raise LAI more greatly in the IR 64 background and therefore NIL-SPIKE may produce higher biomass than IR 64. Taken together, our results indicate that the Daringan SPIKE allele and the Koshihikari GPS allele may not play a role in increasing yield potential at least under the present field conditions, and that other genetic factors that increase sink capacity or biomass production are necessary to improve yield potential in Takanari and IR 64. Interestingly, Takanari yielded significantly greater than IR 64 despite the similar biomass production. Takanari produced more spikelets per unit biomass than IR 64 (Figure ). The higher efficiency of spikelet production per unit biomass in Takanari may be due to the alleles of GN1a and APO1 that increase spikelet number per panicle (Takai et al., Citation2014). We are currently conducting genetic analysis using materials derived from a cross between Takanari and IR 64 to understand the genetic mechanisms underlying the higher efficiency of spikelet production per unit biomass and to clarify whether the genetic factors in Takanari can improve yield potential in IR 64.
Conclusion
Although two SNPs were revealed in the promoter region between Koshihikari and Daringan in the NAL1 gene (the causal gene of both SPIKE and GPS), the effects of both alleles were similar but were affected by the difference of the genetic backgrounds. The environment factors such as different vegetative growth period and soil fertility conditions may also influence the effect of SPIKE/GPS on yield potential. The interaction between QTLs and environment should be important to utilize them to breeding programs and to farmers. Further yield trials across multiple environments (as well as cultivation method improvement that can compensate for the negative environmental effects) will confirm the utility of SPIKE/GPS.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was supported by the NARO Institute of Crop Science, Japan.
Acknowledgements
We thank the staff of the research support section at NARO Agricultural Research Center.
References
- De Datta, S. K., Tauro, A. C., & Balaoing, S. N. (1968). Effect of plant type and nitrogen level on the growth characteristics and grain yield of indica rice in the tropics. Agronomy Journal, 60, 643–647.10.2134/agronj1968.00021962006000060017x
- Dingkuhn, M., Laza, R. C., Kumar, U., Mendez, K. S., Collard, B., Jagadish, K., … Sow, A. (2015). Improving yield potential of tropical rice: Achieved levels and perspectives through improved ideotypes. Field Crops Research, 182, 43–59.10.1016/j.fcr.2015.05.025
- Fujita, D., Trijatmiko, K. R., Tagle, A. G., Sapasap, M. V., Koide, Y., Sasaki, K., … Kobayashi, N. (2013). NAL1 allele from a rice landrace greatly increases yield in modern indica cultivars. Proceedings of the National Academy of Sciences of the United States of America, 110, 20431–20436.10.1073/pnas.1310790110
- Imbe, T., Akama, Y., Nakane, A., Hata, T., Ise, K., Ando, I., … Koga, Y. (2004). Development of a multipurpose high-yielding rice variety ‘Takanari’. Bulletin of the National Institute of Crop Science, 5, 35–51. In Japanese with English summary.
- Khush, G. S. (2001). Green revolution: The way forward. Nature Reviews Genetics, 2, 815–822.10.1038/35093585
- Peng, S., Cassman, K. G., Virmani, S. S., Sheehy, J., & Khush, G. S. (1999). Yield potential trends of tropical rice since the release of IR 8 and the challenge of increasing rice yield potential. Crop Science, 39, 1552–1559.10.2135/cropsci1999.3961552x
- Peng, S., Khush, G. S., Virk, P., Tang, Q., & Zou, Y. (2008). Progress in ideotype breeding to increase rice yield potential. Field Crops Research, 108, 32–38.10.1016/j.fcr.2008.04.001
- Peng, S., Laza, R. C., Visperas, R. M., Sanico, A. L., Cassman, K. G., & Khush, G. S. (2000). Grain yield of rice cultivars and lines developed in the Philippines since 1966. Crop Science, 40, 307–314.10.2135/cropsci2000.402307x
- Qi, J., Qian, Q., Bu, Q., Li, S., Chen, Q., Sun, J., … Li, C. (2008). Mutation of the rice narrow leaf1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiology, 147, 1947–1959.10.1104/pp.108.118778
- Taguchi-Shiobara, F., Ota, T., Ebana, K., Ookawa, T., Yamasaki, M., Tanabata, T., … Yano, M. (2015). Natural variation in the flag leaf morphology of rice due to a mutation of the narrow leaf 1 gene in Oryza sativa L. Genetics, 201, 795–808.10.1534/genetics.115.181040
- Takai, T., Adachi, S., Taguchi-Shiobara, F., Sanoh-Arai, Y., Iwasawa, N., Yoshinaga, S., … Yamamoto, T. (2013). A natural variant of NAL1, selected in high-yield rice breeding programs, pleiotropically increases photosynthesis rate. Scientific Reports, 3, 2149.10.1038/srep02149
- Takai, T., Ikka, T., Kondo, K., Nonoue, Y., Ono, N., Arai-Sanoh, Y., … Yamamoto, T. (2014). Genetic mechanisms underlying yield potential in the rice high-yielding cultivar Takanari, based on reciprocal chromosome segment substitution lines. BMC Plant Biology, 14, 295.10.1186/s12870-014-0295-2
- Takai, T., Matsuura, S., Nishio, T., Ohsumi, A., Shiraiwa, T., & Horie, T. (2006). Rice yield potential is closely related to crop growth rate during late reproductive period. Field Crops Research, 96, 328–335.10.1016/j.fcr.2005.08.001
- Takai, T., Yano, M., & Yamamoto, T. (2009). Canopy temperature on clear and cloudy days can be used to estimate varietal differences in stomatal conductance in rice. Field Crops Research, 115, 165–170.
- Takeuchi, Y., Hori, K., Suzuki, K., Nonoue, Y., Takemoto-Kuno, Y., Maeda, H. … Ando, I. (2008). Major QTLs for eating quality of an elite Japanese rice cultivar, Koshihikari, on the short arm of chromosome 3. Breeding Science, 58, 437–445.10.1270/jsbbs.58.437
- Tester, M., & Langridge, P. (2010). Breeding technologies to increase crop production in a changing world. Science, 327, 818–822.10.1126/science.1183700
- The 3000 rice genomes project. (2014). The 3000 rice genomes project. Gigascience, 3, 7.
- Uga, Y., Okuno, K., & Yano, M. (2011). Dro1, a major QTL involved in deep rooting of rice under upland field conditions. Journal of Experimental Botany, 62, 2485–2494.10.1093/jxb/erq429