ABSTRACT
Low oxygen concentrations in flooded paddy fields cause poor seedling establishment in wet direct seeded rice. We aimed to identify novel genomic regions associated with coleoptile elongation under anaerobic conditions in order to improve seedling establishment. Chromosome segment substitution lines (CSSLs), substituted with Koshihikari (Japonica-type) in the IR 64 genetic background (Indica-type) were evaluated. These lines were imbibed with a hydroponic solution containing Oxyrase to create a stable anaerobic condition and coleoptile lengths were measured six days after incubation at 30°C. Among the CSSLs, only SL2111 had a significantly longer coleoptile (23.9 mm) than that of IR 64 (14.3 mm). From genotype and phenotype analyses, a novel QTL, referred to as qACE3.1, for coleoptile elongation under anaerobic conditions was detected on chromosome 3. To explore the molecular mechanism of qACE3.1, the expression levels of genes encoding enzymes involved in starch degradation and fermentation were assessed. In SL2111 coleoptiles, the expression levels of pyruvate decarboxylase were significantly higher than in IR 64 coleoptiles whereas those of alcohol dehydrogenase were lower. In contrast, no differences were observed in the expression levels of genes associated with starch degradation. These results imply that qACE3.1 may specifically affect fermentative metabolism. In addition, we evaluated the impact of qACE3.1 on seedling establishment under flooded conditions in a paddy field.
GRAPHICAL ABSTRACT
Introduction
Direct seeded rice (DSR) helps meet the challenges caused by water and labor shortages, and time and cropping area conflicts. DSR is being adopted in both rainfed and irrigated lowland ecosystems. In spite of these advantages, spread of DSR is limited by its lower yield because of poor seedling establishment, especially with wet DSR. In rainfed areas where DSR is practiced, farmers often encounter flooding and/or waterlogging when it rains immediately after seeding, which leads to a severe reduction in, or complete failure of, seedling establishment because of the sensitivity of rice to low oxygen stress caused by flooding during germination (Miro & Ismail, Citation2013). Soil waterlogging or flooding also occurs when the land is not well-levelled in irrigated areas (Kirk et al., Citation2014). These problems of poor seedling emergence and establishment in flooded soils is further compounded by the subsequent invasion of weeds (Ismail et al., Citation2012). To overcome these problems, developing of new varieties can emerge their shoot from flooded soils would be beneficial way for DSR in these areas (Ismail et al., Citation2012). Thus, it is important to develop breeding materials that can withstand flooding during seedling emergence.
To develop improved breeding materials for seedling establishment under flooded conditions in DSR, quantitative trait loci (QTLs) for the rate of seedling emergence under anaerobic conditions (i.e., anaerobic germination) have been determined (Angaji, Septiningsih, Mackill & Ismail, Citation2010; Baltazar et al., Citation2014, Citation2019; Kim & Reinke, Citation2018; Septiningsih et al., Citation2013). Among these QTLs, a trehalose-6-phosphate phosphatase gene (OsTPP7) was identified as the gene responsible for qAG-9-2 a major quantitative trait locus (QTL) for anaerobic germination (Kretzschmar et al., Citation2015). OsTPP7 is considered to be associated with starch mobilization which drives the growth kinetics of the germinating embryo and elongating the coleoptile under anaerobic conditions. In addition, a near isogenic line of qAG-9-2 showed better seedling establishment under flooding stress compared with the background variety, showing a higher grain yield in the paddy field (Chamara et al., Citation2018; Lal et al., Citation2018). In high latitude areas such as in East Asia, seedling emergence were observed under low temperature conditions in addition to flooding stress (Fukuda et al., Citation2014; Iwata et al., Citation2010). Coleoptile elongation under anaerobic conditions is considered to be one of the most important traits associated with seedling establishment (Adachi et al., Citation2015; Ogiwara & Terashima, Citation2009). Coleoptiles from tolerant cultivars grow faster and longer under submerged soil conditions, and this morphological adaptation enables them to reach surface O2 faster, allowing diffusion through this structure to other organs including the primary leaf and root to support seedling growth (Hsu & Tung, Citation2017). Many genetic loci associated with coleoptile elongation under anaerobic conditions were found by genome wide association studies (GWAS) (Hsu & Tung, Citation2015; Nghi et al., Citation2019; Zhang et al., Citation2017) and QTL analysis (Jiang et al., Citation2006; Kuya et al., Citation2019; Manangkil et al., Citation2013). However, only a few QTLs have been characterized using substitution lines or near isogenic lines to assess their impact on seedling establishment in the paddy field and few have addressed the molecular mechanisms for tolerance to anaerobic conditions.
In addition to the ability to detect genetic loci responsible for tolerance to anaerobic conditions, physiological analyses have also been performed to reveal mechanisms that provide tolerance to anaerobic conditions. (Ismail et al., Citation2009). In a comprehensive analysis of transcript and proteomic profiles, genes involved in starch degradation and fermentation were found to be upregulated in O2-deficient rice seedlings (Hsu & Tung, Citation2017; Lasanthi-Kudahettige et al., Citation2007). When the oxygen concentration is very low, carbohydrate metabolism is strongly inhibited, especially the steps involved in the breakdown of starch into simple sugars for use in glycolysis (Miro & Ismail, Citation2013). Among the enzymes involved in sugar degradation, amylase activity increases during germination and coleoptile elongation under anaerobic conditions. Maintenance of higher α-amylase activity has been widely reported to be an important step in carbohydrate catabolism under conditions of low oxygen concentration (Lasanthi-Kudahettige et al., Citation2007), and a higher activity is observed in the seeds of tolerant genotypes germinating under flooding condition (Ismail et al., Citation2009). Among the genes encoding α-amylases, RAmy3D expression is positively correlated with coleoptile elongation and seedling survival, especially in tolerant genotypes (Ismail et al., Citation2009). Fermentative metabolism or anaerobic respiration in submerged coleoptiles uses the alcohol, lactate, and alanine fermentation pathways to regenerate the NAD+ required for glycolysis. Among these three main pathways, alcoholic fermentation is considered the most important, since about 92% of the pyruvate generated through glycolysis is directed to ethanol production, and only 7% to the lactate pathway and 1% to the alanine pathway (Kato-Noguchi, Citation2006). Alcoholic fermentation is strongly activated in rice during germination under conditions of oxygen deprivation, as reflected in the induction of the key enzymes, including pyruvate decarboxylase (PDC), alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH) (Hsu & Tung, Citation2017; Miro et al., Citation2017). PDC channels pyruvate into the alcohol fermentation pathway and catalyzes the decarboxylation of pyruvate to acetaldehyde (Kato-Noguchi, Citation2006). The acetaldehyde produced is metabolized in one of two ways; either by conversion to ethanol by ADH or conversion to acetate by ALDH (Kato-Noguchi, Citation2006). The expression levels of the mRNAs encoding these enzymes and the protein and enzymatic activity are also up regulated in a tolerant variety under anaerobic conditions (Hsu & Tung, Citation2017; Miro et al., Citation2017). Therefore, when QTLs for seedling establishment are found, characterizing these enzymes allow us to understand the mechanisms by which the QTLs function in seedling establishment.
Here, we aimed to detect novel QTLs for coleoptile elongation under anaerobic condition by using oxyrase which consumes dissolved oxygen in the hydroponic solution. It has been observed that there is a high correlation between seedling establishment in the paddy field and coleoptile length under hydroponic culture conditions in which oxygen levels were reduced by oxyrase (Ogiwara & Terashima, Citation2007). We utilized this method to identify QTLs for coleoptile elongation under anaerobic conditions because the method has never been used to evaluate populations for genetic analysis even though it can easily reduce oxygen levels. We evaluated chromosome segment substitution lines (CSSLs) in the IR 64 genetic background (short coleoptiles) crossed with Koshihikari (long coleoptiles) as the donor parent (Nagata et al., Citation2015; Ujiie et al., Citation2016). Indica-type rice varieties such as IR 64 are grown predominantly in southern China and South and Southeast Asia (Mackill & Khush, Citation2018; Xie & Zhang, Citation2018). As IR 64 has broad adaptability, early maturity, and high quality, it has been released in many countries and has become a popular variety. It is also used extensively as a parent in breeding programs, and to develop populations for genetic analysis (Mackill & Khush, Citation2018). By using selected CSSLs, we evaluated the impact of the QTL detected here on seedling establishment in the paddy field and physiological function during coleoptile elongation.
Materials and methods
Plant materials
Chromosome segment substitution lines (CSSLs) and their genotypes were provided by the Rice Genome Resource Center in the National Agriculture and Food Research Organization (http://www.rgrc.dna.affrc.go.jp/index.html). For the QTL analysis, 39 CSSLs which have part of their chromosome segment substituted with Koshihikari in the IR 64 genetic background were used except for SL2119 whose seeds were not obtained due to late heading and sterility in the Kanto region of Japan. For field evaluation of seedling establishment, SL2008 which was substituted with IR 64 on chromosome 3 on the Koshihikari genetic background was used. In 2015, CSSLs were cultivated to obtain healthy seeds in an experimental paddy field in the Institute for Sustainable Agro‑ecosystem Services, Graduate School of Agricultural and Life Sciences, The University of Tokyo (35°74′N, 139°54′E). Seeds were harvested approximately 40 days after heading, dried for more than a month in a well-ventilated greenhouse, and subsequently stored below 10°C in an air-tight box containing a desiccant to maintain seed viability. Seed dormancy was broken by drying for 10 days at 40°C prior to using for experimentation performed in 2016.
Evaluation of coleoptile elongation and QTL detection
Seeds were submerged in hot water at 55°C for 10 min to sterilize them as described in Masuda et al. (Citation2018). The sterilized seeds were placed in glass petri dishes with 0.2% Plant Preservative Mixture™ (PPM-100, Plant Cell Technology supplied by Nakarai Tesc Co., Kyoto, Japan) solution and incubated at 15°C for four days. One day before sowing, the temperature of the incubation was increased to 30°C to synchronize the seeds. To create a severe anaerobic condition in hydroponic solution, Oxyrase (Oxyrase Inc. Mansfield, Ohio) were used in the previous study (Ogiwara & Terashima, Citation2007). In the present study, we added a 1/500 volume of oxyrase to an autoclaved incubation solution comprising Murashige and Skoog (MS) medium diluted 20 fold. During coleoptile elongation, the dissolved oxygen concentration of the incubation solution was below 2 mg L−1, as determined using a Dissolved Oxygen Meter (DO-31P; DKK-TOA Co., Tokyo). A well-imbibed seed was put into the bottom of a glass tube (φ13 mm, 10 mL volume) filled with 4 mL of MS medium. The glass tubes were wrapped with black cloth and put in an incubator. After 6 days incubation at 30°C under dark conditions, the coleoptile lengths of 15 plants in each line were measured. Significant differences in coleoptile length between CSSLs and IR 64 were assessed using Dunnett’s test at a 1% significance level. The effects of a Koshihikari substituted segment are shown as chromosome regions affecting a trait (CRATs) as described in Ujiie et al. (Citation2016).
Gene expression analysis
Coleoptiles and embryos were obtained using the procedure described in Evaluation of coleoptile elongation and QTL detection, with minor modifications. As alternatives to imbibed seeds, we used sterilized dry seeds for gene expression analysis and field experiments. Seven days after incubation at 30°C, we used replicates of five plants for the extraction of total RNA. Coleoptiles and embryos were divided with a knife from the endosperm and placed in liquid nitrogen and then stored at −80°C until total RNA extraction. Total RNA was extracted from the coleoptiles and embryos with an RNeasy Mini Kit (Qiagen, Hilden, Germany). The total extracted RNA was incubated with DNase (Recombinant DNase I, TaKaRa Bio Inc., Shiga, Japan) to digest any contaminating DNA. After inactivating the DNase, 1.5 μg of total RNA was used for the reverse transcription reaction using PrimeScript™ RT Master Mix (TaKaRa Bio Inc., Shiga, Japan) to make single stranded DNA. For real time RT-PCR, PCR was performed with a KOD SYBR® qPCR Mix (Toyobo Co, Osaka, Japan) using CFX Connect™ (Bio-Rad, Hercules, CA, USA). The primer sequences used for the gene expression analysis are listed in Table S1. These target genes are not located in the qACE3.1 candidate region. Differences in gene expression between IR 64 and SL2111 were compared using the ∆∆Ct method (Livak & Schmittgen, Citation2001), with significant differences being determined using a t-test (n = 4). UBQ5 was used as the control gene.
Field experiments
Evaluation of SL2111 seedling establishment was performed in 2018 under flooding conditions in soil from the same experimental paddy field described in Plant Materials. To estimate the effect of the IR 64 allele in qACE3.1 on the genetic background in Koshihikari, SL2008 which has a substitution on chromosome 3 including the qACE3.1 candidate region (Figure S1) was used for this field experiment. A total of 50 dry seeds each from IR 64, Koshihikari, SL2111, and SL2008 were seeded into a cell-tray (Minoru 448 nursery box, Minoru, Okayama) filled with puddled soil from the paddy field. One seed was sown in each cell (φ16 mm, 25 mm depth) at a depth of 10 mm. The four lines were located in one tray with a randomized block deign that included five replicates. These trays were then put on a paddled paddy field. The water depth was kept at 7 ± 1 cm. The number of established seedlings were counted at 35 days after seeding, and sampled from 50 cells in each replication for measurement of dry weight of seedlings.
Results
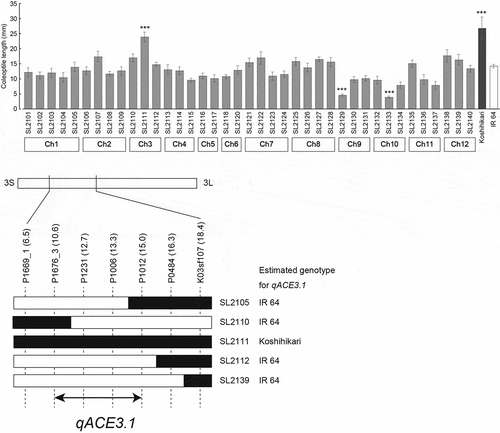
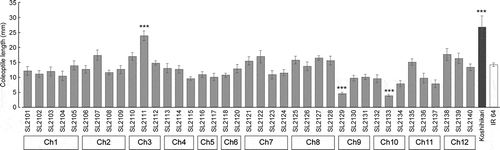
CSSLs and their parental cultivars were incubated at 30°C for six days in an anaerobic MS solution that had a reduced dissolved oxygen concentration created using Oxyrase. The coleoptile length of Koshihikari (26.8 mm) was significantly higher than that of IR 64 (14.3 mm) (). Among the CSSLs, only SL2111 showed a significantly longer coleoptile length (23.9 mm) than that of IR 64. SL2129 and SL2133 had significantly shorter coleoptile lengths than that of IR 64. Based on the result from the genotype and phenotype analyses of CSSLs, the CRATs were determined. The effect and position of the CRATs are summarized in . Among these CRATs, only the CRAT on chromosome 3 improved the coleoptile length of IR 64 under anaerobic conditions. We have named this QTL qACE3.1 (anaerobic coleoptile elongation) and mapped it to the region between P1676_3 and P1012 ().
Table 1. Effects of CRATs for coleoptile elongation for Koshihikari alleles in the IR 64 genetic background.
Figure 1. Coleoptile length of chromosome segment substitution lines grown under low oxygen conditions.
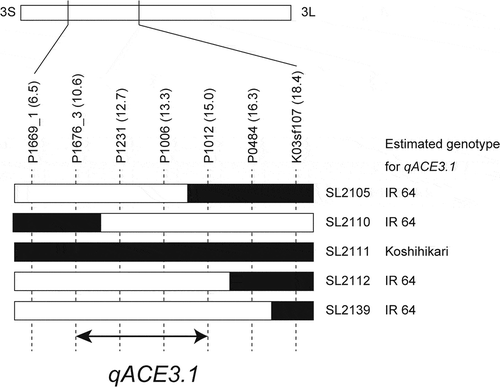
Figure 2. The candidate qACE3.1 region on chromosome 3 for coleoptile elongation under anaerobic conditions.
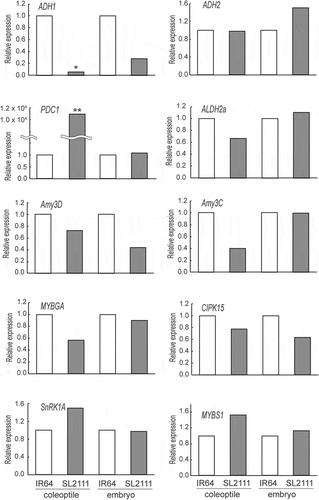
To elucidate the mechanism of the effect of qACE3.1, gene expression levels in the coleoptiles and embryos from IR 64 and SL2111 seeds grown under anaerobic conditions were evaluated. Because sugar metabolism and fermentation are known to be important for coleoptile elongation, we selected genes that encoded enzymes such as amylase (Amy3D and Amy3 C) (Lee et al., Citation2014), alcohol dehydrogenase (ADH1 and 2) (Saika et al., Citation2006), acetaldehyde dehydrogenase (ALDH2a) (Miro & Ismail, Citation2013), pyruvate decarboxylase (PDC1)(Kato-Noguchi, Citation2006). In addition, transcription factors which are involved in controlling amylase gene expression such as MYBS1 (Lu et al., Citation2002), MYBGA (Chen et al., Citation2006), SnRK1A (Lu et al., Citation2007), and CIPK15 (Lee et al., Citation2009) were also analyzed. Among these genes, only the expression of PDC1 in the SL2111 coleoptile was significantly higher than that of IR 64 (). In contrast, ADH1 gene expression was significant lower in the SL2111 coleoptile (). Lower gene expression of ADH1 was also observed in the embryo although there were no significant differences in ADH1 expression between two lines (). The expression levels of other genes such as ADH2 and ALDH2a in SL2111 were found to be the same as IR 64. A lower gene expression level of amylase genes, especially Amy3D, was observed in SL2111 even though this was not significant. There were no trends in the expression levels of genes involved in controlling amylase gene expression.
Figure 3. Comparison of gene expression levels associated with anaerobic coleoptile elongation between IR 64 and SL2111 at seven days after incubation.
To evaluate the effect of qACE3.1 on seedling establishment in the paddy field, SL2111 and SL2008 were used for a field trial. In SL2008, chromosome segment including the qACE3.1 candidate region on chromosome 3 was substituted with IR 64 in the Koshihikari genetic background. We hypothesized that the IR 64 allele of qACE3.1 would have a reduced rate of seedling emergence in the Koshihikari background. The effect of allelic differences and background genotype were significant (). The ratio of seedling emergence (37.6%) and seedling dry weight (2.5 g) in plants harboring the Koshihikari allele were both higher than those in plants containing the IR 64 allele (27.6% and 1.7 g, respectively). Significant differences were also observed in the ratio of seedling emergence between Koshihikari and SL2008 lines, although we detected no significant different in the seedling emergence of IR 64 and SL2111 plants ().
Table 2. Seedling establishment of chromosome segment substitution lines in paddy fields under flooded conditions.
Discussion
We evaluated coleoptile elongation under anaerobic conditions by adding the enzyme Oxyrase into the hydroponic solution to reduce the oxygen concentration. qACE3.1 was successfully detected on the short arm of chromosome 3 in the population of CSSLs which have part of their chromosome segment substituted with Koshihikari in the IR 64 genetic background. In previous studies, many QTLs for seedling establishment have been detected in the Indica-type genetic background to improve adaptability to the wet direct seeding method (Angaji et al., Citation2010; Baltazar et al., Citation2014, Citation2019; Kim & Reinke, Citation2018; Septiningsih et al., Citation2013). In addition, two genetic loci for coleoptile elongation under anaerobic conditions have been detected by GWAS (Hsu & Tung, Citation2015; Nghi et al., Citation2019) and QTL analyses (Jiang et al., Citation2006; Manangkil et al., Citation2013). Among these QTLs for seedling establishment, three have been detected on chromosome 3 in populations derived from cross combinations between IR 64 and Khao Hlan On (Angaji et al., Citation2010), IR 64 and Kharsu 80A (Baltazar et al., Citation2019), and IR 64 and Nanhi (Baltazar et al., Citation2014), which were mapped to regions at approximately 30.9, 5.5, and 21.1 Mb, respectively, on chromosome 3. Moreover, using GWAS, a further QTL has been detected on chromosome 3 in a Japonica-type population, which is located at approximately 27.4 Mb (Nghi et al., Citation2019). However, the qACE3.1 detected in the present study is located in the genomic region 10.6 to 15.0 Mb on chromosome 3. qACE3.1 therefore appears to be a different QTL from the previously detected QTLs for seedling establishment on chromosome 3. Therefore, we consider qACE3.1 to be a novel QTL for coleoptile elongation under anaerobic conditions. Further fine mapping is needed to identify the responsible gene(s) in qACE3.1 and to developed tightly linked DNA markers for marker assisted selection. Recently, a QTL on chromosome 9 for anaerobic germination, which may be identical to qAG-9-2, was detected in the same CSSL population under more severer oxygen concentration (<0.2 mg L−1) than the present study (Kuya et al., Citation2019). These results imply that detected QTLs may respond to the different oxygen concentration during germination and subsequent coleoptile elongation. We believe that the differences between the present and previous studies regarding QTL detection can largely be attributed to differences in the respective methodologies. By subjecting seedlings to alternative conditions such as oxygen concentration, we suspect that other QTLs would be detected and that this would subsequently contribute to the development of lines with enhanced seedling emergence, which in turn would provide desirable materials for developing pyramiding lines in varieties with an Indica-type genetic background.
To elucidate part of the molecular mechanism of qACE3.1 in SL2111, we conducted a gene expression analysis. We targeted genes which were associated with coleoptile elongation under anaerobic conditions. Among these genes, the expression levels of the PDC1 gene were dramatically up-regulated in SL2111 carrying qACE3.1 compared with IR 64 (). PDC catalyzes the decarboxylation of pyruvate to acetaldehyde, and is activated under anaerobic conditions during coleoptile elongation (Kato-Noguchi, Citation2006). The enzymatic activity of PDC is higher in cultivars tolerant to anoxia compared to sensitive cultivars (Ismail et al., Citation2009). Moreover, the rice cultivar ‘Taipei 309’ which over-expresses PDC1 gene has a higher tolerance to anoxia due to an increase in alcohol metabolism (Quimio et al., Citation2000). Therefore, increasing the gene expression levels of PDC1 may contribute to coleoptile elongation in SL2111 under anaerobic conditions. These results and the previous reports suggest that enzymes such as ADH and ALDH that are capable of metabolizing acetaldehyde may be activated. Acetaldehyde is metabolized in one of two ways; conversion to ethanol by ADH or conversion to acetate by ALDH. ADH quickly metabolizes acetaldehyde to reduce the risk of acetaldehyde-mediated cytotoxicity, and to generate NAD+ for glycolysis, we found that the expression levels of ADH1, which encodes ADH, were significantly lower in SL2111 than in IR 64. At present, however, we are unable to explain why SL2111 shows lower ADH1 expression, even though this line was characterized by a longer coleoptile under anaerobic conditions. In this regard, a recent study in Arabidopsis revealed that ADH activity was up-regulated in a PDC-inactivated mutant (Bui et al., Citation2019). Given that the authors observed that gene expression levels in mutant and wild-type plants were essentially the same, they concluded that inactivation of PDC activity promotes the accumulation of ADH protein (Bui et al., Citation2019). We suspect that similar, as yet undetermined, mechanisms may suppress gene expression of ADH in rice during germination. In the alternative pathway associated with aldehyde metabolism, ALDH is also assumed to play an important role in anaerobic germination tolerance, given that it’s activity and gene expression levels are also higher during coleoptile elongation in a rice variety tolerant to anaerobic conditions (Miro & Ismail, Citation2013). Further studies are accordingly needed to examine the changes in metabolic turnover subsequent to the PDC step. Interestingly, the expression of α-amylase genes and the transcription factors which regulate α-amylase genes in SL2111 were found at the same level as in IR 64 which was contrary to our expectations. From these data, qACE3.1 may be associated with the specific regulation of the activities of fermentation enzyme during coleoptile elongation under anaerobic conditions. However, although we detected differences between IR 64 and SL2111 with respect to the coleoptile expression levels of certain genes at 7 days after incubation, in order to clearly establish the function of qACE3.1, it will be necessary to perform analyses for a range of time points. Moreover, comprehensive gene expression analysis will contribute to a more detailed assessment of the difference between IR 64 and SL2111.
Although many QTLs for seedling establishment have been found previously, the impact of a QTL on seedling establishment under field conditions has however, with the exception of qAG-9-2, never been validated (Lal et al., Citation2018). IR64-AG NILs carrying qAG-9-2 had a 217.5% higher plant population compared to IR 64, irrespective of the establishment methods. In the present study, the allelic effect of qACE3.1 on seedling establishment was confirmed under flooded conditions in paddy fields (). Therefore, the Koshihikari allele of qACE3.1 may associate with seedling establishment of Indica-type varieties in a flooded paddy field. However, we also found that SL2111 showed the same level of seedling establishment as IR 64 under field conditions, even though we observed that the coleoptile length of SL2111 seedlings was greater than that of IR 64 in an anaerobic hydroponic solution. In flooded paddy soil, although oxygen deficiency is considered to be main stress factor contributing to poor seedling establishment, other factors such as an increase sulfide ion concentration associated with soil reduction have been identified (Hara, Citation2013). Therefore, in addition to qACE3.1, we suspect that it will be necessary to identify other genetic factors involved in seedling establishment in Koshihikari in the same CSSL population to close the phenotypical gap between SL2111 and Koshihikari for seedling establishment in paddy fields.
PPS2019_109RP-File008.pdf
Download PDF (852.2 KB)PPS2019_109RP-File007.pdf
Download PDF (12.2 KB)Acknowledgments
We thank R. Soga and H. Teshima (The University of Tokyo) for their technical assistance in carrying out experiments.
Disclosure Statement
The authors report that they have no potential conflict of interest.
Supplementary Material
The supplementary data for this article can be accessed here.
Additional information
Funding
References
- Adachi, Y., Sugiyama, M., Sakagami, J.-I., Fukuda, A., Ohe, M., & Watanabe, H. (2015). Seed germination and coleoptile growth of new rice lines adapted to hypoxic conditions. Plant Production Science, 18(4), 471–475. https://doi.org/10.1626/pps.18.471
- Angaji, S. A., Septiningsih, E. M., Mackill, D. J., & Ismail, A. M. (2010). QTLs associated with tolerance of flooding during germination in rice (Oryza sativa L.). Euphytica, 172(2), 159–168. https://doi.org/10.1007/s10681-009-0014-5
- Baltazar, M. D., Ignacio, J. C. I., Thomson, M. J., Ismail, A. M., Mendioro, M. S., & Septiningsih, E. M. (2014). QTL mapping for tolerance of anaerobic germination from IR64 and the aus landrace Nanhi using SNP genotyping. Euphytica, 197(2), 251–260. https://doi.org/10.1007/s10681-014-1064-x
- Baltazar, M. D., Ignacio, J. C. I., Thomson, M. J., Ismail, A. M., Mendioro, M. S., & Septiningsih, E. M. (2019). QTL mapping for tolerance to anaerobic germination in rice from IR64 and the aus landrace Kharsu 80A. Breeding Science, 69(2), 227–233. https://doi.org/10.1270/jsbbs.18159
- Bui, L. T., Novi, G., Lombardi, L., Iannuzzi, C., Rossi, J., Santaniello, A., Mensuali, A., Corbineau, F., Giuntoli, B., Perata, P., Zaffagnini, M., & Licausi, F. (2019). Conservation of ethanol fermentation and its regulation in land plants. Journal of Experimental Botany, 70(6), 1815–1827. https://doi.org/10.1093/jxb/erz052
- Chamara, B. S., Marambe, B., Kumar, V., Ismail, A. M., Septiningsih, E. M., & Chauhan, B. S. (2018). Optimizing sowing and flooding depth for anaerobic germination-tolerant genotypes to enhance crop establishment, early growth, and weed management in dry-seeded rice (Oryza sativa L.). Frontiers in Plant Science, 9, 1–15. https://doi.org/10.3389/fpls.2018.01654
- Chen, P. W., Chiang, C. M., Tseng, T. H., & Yu, S. M. (2006). Interaction between rice mybga and the gibberellin response element controls tissue-specific sugar sensitivity of α-amylase genes. The Plant Cell, 18(9), 2326–2340. https://doi.org/10.1105/tpc.105.038844
- Fukuda, A., Kataoka, K., Shiratsuchi, H., Fukushima, A., Yamaguchi, H., Mochida, H., & Ogiwara, H. (2014). QTLs for seedling growth of direct seeded rice under submerged and low temperature conditions. Plant Production Science, 17(1), 41–46. https://doi.org/10.1626/pps.17.41
- Hara, Y. (2013). Suppressive effect of sulfate on establishment of rice seedlings in submerged soil may be due to sulfide generation around the seeds. Plant Production Science, 16(1), 50–60. https://doi.org/10.1626/pps.16.50
- Hsu, S. K., & Tung, C. W. (2015). Genetic mapping of anaerobic germination-associated qtls controlling coleoptile elongation in rice. Rice, 8(1), 1–12. https://doi.org/10.1186/s12284-015-0072-3
- Hsu, S. K., & Tung, C. W. (2017). RNA-Seq analysis of diverse rice genotypes to identify the genes controlling coleoptile growth during submerged germination. Frontiers in Plant Science, 8, 1–15. https://doi.org/10.3389/fpls.2017.00762
- Ismail, A. M., Ella, E. S., Vergara, G. V., & Mackill, D. J. (2009). Mechanisms associated with tolerance to flooding during germination and early seedling growth in rice (Oryza sativa). Annals of Botany, 103(2), 197–209. https://doi.org/10.1093/aob/mcn211
- Ismail, A. M., Johnson, D. E., Ella, E. S., Vergara, G. V., & Baltazar, A. M. (2012). Adaptation to flooding during emergence and seedling growth in rice and weeds, and implications for crop establishment. AoB PLANTS, 2012, 1–18. https://doi.org/10.1093/aobpla/pls019
- Iwata, N., Shinada, H., Kiuchi, H., Sato, T., & Fujino, K. (2010). Mapping of QTLs controlling seedling establishment using a direct seeding method in rice. Breeding Science, 60(4), 353–360. https://doi.org/10.1270/jsbbs.60.353
- Jiang, L., Liu, S., Hou, M., Tang, J., Chen, L., Zhai, H., & Wan, J. (2006). Analysis of QTLs for seed low temperature germinability and anoxia germinability in rice (Oryza sativa L.). Field Crops Research, 98(1), 68–75. https://doi.org/10.1016/j.fcr.2005.12.015
- Kato-Noguchi, H. (2006). Pyruvate metabolism in rice coleoptiles under anaerobiosis. Plant Growth Regulation, 50(1), 41–46. https://doi.org/10.1007/s10725-006-9124-4
- Kim, S.-M., & Reinke, R. F. (2018). Identification of QTLs for tolerance to hypoxia during germination in rice. Euphytica, 214(9), 160. https://doi.org/10.1007/s10681-018-2238-8
- Kirk, G. J. D., Greenway, H., Atwell, B. J., Ismail, A. M., & Colmer, T. D. (2014). Adaptation of rice to flooded soils. In Progress in Botany, 75, 215–253. https://doi.org/10.1007/978-3-642-38797-5_8
- Kretzschmar, T., Pelayo, M. A. F., Trijatmiko, K. R., Gabunada, L. F. M., Alam, R., Jimenez, R., Mendioro, M. S., Slamet-Loedin, I. H., Sreenivasulu, N., Bailey-Serres, J., Ismail, A. M., Mackill, D. J., & Septiningsih, E. M. (2015). A trehalose-6-phosphate phosphatase enhances anaerobic germination tolerance in rice. Nature Plants, 1(9), 15124. https://doi.org/10.1038/nplants.2015.124
- Kuya, N., Sun, J., Iijima, K., Venuprasad, R., & Yamamoto, T. (2019). Novel method for evaluation of anaerobic germination in rice and its application to diverse genetic collections. Breeding Science, 69(4), 633–639. https://doi.org/10.1270/jsbbs.19003
- Lal, B., Gautam, P., Nayak, A. K., Raja, R., Shahid, M., Tripathi, R., Singh, S., Septiningsih, E. M., & Ismail, A. M. (2018). Agronomic manipulations can enhance the productivity of anaerobic tolerant rice sown in flooded soils in rainfed areas. Field Crops Research, 220, 105–116. https://doi.org/10.1016/j.fcr.2016.08.026
- Lasanthi-Kudahettige, R., Magneschi, L., Loreti, E., Gonzali, S., Licausi, F., Novi, G., Beretta, O., Vitulli, F., Alpi, A., & Perata, P. (2007). Transcript profiling of the anoxic rice coleoptile. Plant Physiology, 144(1), 218–231. https://doi.org/10.1104/pp.106.093997
- Lee, K. W., Chen, P. W., Lu, C. A., Chen, S., Ho, T. H. D., & Yu, S. M. (2009). Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Science Signaling, 2(91), ra61. https://doi.org/10.1126/scisignal.2000333
- Lee, K. W., Chen, P. W., & Yu, S. M. (2014). Metabolic adaptation to sugar/O2 deficiency for anaerobic germination and seedling growth in rice. Plant, Cell & Environment, 37(10), 2234–2244. https://doi.org/10.1111/pce.12311
- Livak, K. J., & Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods, 25(4), 402–408. https://doi.org/10.1006/meth.2001.1262
- Lu, C. A., Ho, T. D., Ho, S. L., & Yu, S. M. (2002). Three novel myb proteins with one DNA binding repeat mediate sugar and hormone regulation of α-amylase gene expression. The Plant Cell, 14(8), 1963–1980. https://doi.org/10.1105/tpc.001735
- Lu, C. A., Lin, C. C., Lee, K. W., Chen, J. L., Huang, L. F., Ho, S. L., Liu, H.-J., Hsing, Y.-I., & Yu, S. M. (2007). The SnRK1A protein kinase plays a key role in sugar signaling during germination and seedling growth of rice. The Plant Cell, 19(8), 2484–2499. https://doi.org/10.1105/tpc.105.037887
- Mackill, D. J., & Khush, G. S. (2018). IR64: A high-quality and high-yielding mega variety. Rice, 11(1), 18. https://doi.org/10.1186/s12284-018-0208-3
- Manangkil, O. E., Vu, H. T. T., Mori, N., Yoshida, S., & Nakamura, C. (2013). Mapping of quantitative trait loci controlling seedling vigor in rice (Oryza sativa L.) under submergence. Euphytica, 192(1), 63–75. https://doi.org/10.1007/s10681-012-0857-z
- Masuda, S., Sasaki, K., Kazama, Y., Kisara, C., Takeda, S., Hanzawa, E., Minamisawa, K., & Sato, T. (2018). Mapping of quantitative trait loci related to primary rice root growth as a response to inoculation with Azospirillum sp. strain B510. Communicative & Integrative Biology, 11(3), 1–6. https://doi.org/10.1080/19420889.2018.1502586
- Miro, B., & Ismail, A. M. (2013). Tolerance of anaerobic conditions caused by flooding during germination and early growth in rice (Oryza sativa L.). Frontiers in Plant Science, 4, 215–253. https://doi.org/10.3389/fpls.2013.00269
- Miro, B., Longkumer, T., Entila, F. D., Kohli, A., & Ismail, A. M. (2017). Rice seed germination underwater: Morpho-physiological responses and the bases of differential expression of alcoholic fermentation enzymes. Frontiers in Plant Science, 8, 1–17. https://doi.org/10.3389/fpls.2017.01857
- Nagata, K., Ando, T., Nonoue, Y., Mizubayashi, T., Kitazawa, N., Shomura, A., Ogiso-Tanaka, E., Mizobuchi, R., Shibaya, T., Ogiso-Tanaka, E., Hori, K., Yano, M., Fukuoka, S., & Matsubara, K. (2015). Advanced backcross QTL analysis reveals complicated genetic control of rice grain shape in a japonica × indica cross. Breeding Science, 65(4), 308–318. https://doi.org/10.1270/jsbbs.65.308
- Nghi, K. N., Tondelli, A., Valè, G., Tagliani, A., Marè, C., Perata, P., & Pucciariello, C. (2019). Dissection of coleoptile elongation in japonica rice under submergence through integrated genome‐wide association mapping and transcriptional analyses. Plant, Cell & Environment, 42(6), 1832–1846. https://doi.org/10.1111/pce.v42.6
- Ogiwara, H., & Terashima, K. (2007). [A novel method for estimating seedling establishment traits of rice (Oryza sativa L.) under reduction]. Japanese Journal of Crop Science, 10, 214. In Japanse with English abstract https://doi.org/10.14829/jcsproc.224.0.214.0
- Ogiwara, H., & Terashima, K. (2009). A varietal difference in coleoptile growth is correlated with seedling establishment of direct seeded rice in submerged field under low-temperature conditions. Plant Production Science, 4(3), 166–172. https://doi.org/10.1626/pps.4.166
- Quimio, C. A., Torrizo, L. B., Setter, T. L., Ellis, M., Grover, A., Abrigo, E. M., Oliva, N. P., Ella, E. S., Carpena, A. L., Ito, O., Peacock, W. J., Dennis, E., & Dattal, S. K. (2000). Enhancement of submergence tolerance in transgenic rice overproducing pyruvate decarboxylase. Journal of Plant Physiology, 156(4), 516–521. https://doi.org/10.1016/S0176-1617(00)80167-4
- Saika, H., Matsumura, H., Takano, T., Tsutsumi, N., & Nakazono, M. (2006). A point mutation of adh1 gene is involved in the repression of coleoptile elongation under submergence in rice. Breeding Science, 56(1), 69–74. https://doi.org/10.1270/jsbbs.56.69
- Septiningsih, E. M., Ignacio, J. C. I., Sendon, P. M. D., Sanchez, D. L., Ismail, A. M., & Mackill, D. J. (2013). QTL mapping and confirmation for tolerance of anaerobic conditions during germination derived from the rice landrace Ma-Zhan Red. Theoretical and Applied Genetics, 126(5), 1357–1366. https://doi.org/10.1007/s00122-013-2057-1
- Ujiie, K., Yamamoto, T., Yano, M., & Ishimaru, K. (2016). Genetic factors determining varietal differences in characters affecting yield between two rice (Oryza sativa L.) varieties, Koshihikari and IR64. Genetic Resources and Crop Evolution, 63(1), 97–123. https://doi.org/10.1007/s10722-015-0237-3
- Xie, F., & Zhang, J. (2018). Shanyou 63: An elite mega rice hybrid in China. Rice, 11(1), 17. https://doi.org/10.1186/s12284-018-0210-9
- Zhang, M., Lu, Q., Niu, X., Niu, X., Wang, C., Feng, Y., Wei, X., Prasad, S. M., Dubey, N. K., Pandey, A. C., Sahi, S., Chauhan, D. K., & Singh, V. P. (2017). Association mapping reveals novel genetic loci contributing to flooding tolerance during germination in indica rice. Frontiers in Plant Science, 8, 1–9. https://doi.org/10.3389/fpls.2017.00001