ABSTRACT
Isolation of an adequate quantity of high-quality RNA is a crucial step in gene expression analysis. Difficulties in sampling and RNA extraction of field-grown crops, especially from roots, are substantial obstacles to understanding actual gene expression in the field. We examined the effectiveness of the RNA extraction method (FRP) using pot- and field-grown soybean roots. High quantity and quality of total RNA could extract by the FRP method from both pot- and field-grown soybean roots at different growth stages. To determine the influence of root washing during the sampling process, roots were washed with water for 5 s, 1 min, and 5 min, then analyzed for aquaporin gene expression levels. Root washing for 5 min affected gene expression of some aquaporins. To obtain RNA that reflects the correct gene expression profile, it is necessary to minimize the effects of the sampling process.
Graphical abstract
KEYWORDS:
Introduction
Recent advances in molecular biology have contributed to the understanding of biological phenomena by evaluating the fundamental processes of gene functions. However, there is limited knowledge regarding gene behavior under natural or agricultural field conditions, because most gene studies are laboratory-based. Under field conditions, because the plants are exposed to various environmental factors, such as temperature, humidity, and solar radiation, the behavior of plant genes under field conditions varies depending on these complex environmental characteristics and this causes difficulty in analyzing gene expression (Nagano et al., Citation2012). Understanding the function of genes related to yield and various stress tolerances under field conditions is important for crop improvement under ongoing climate change. Therefore, it is important to understand the behavior of genes under field conditions for gene expression analysis.
Gene expression analysis requires the extraction of high-quality RNA. Many RNA extraction methods for various plant species have been developed; however, difficulty in RNA extraction from belowground parts, such as rhizomes, wooden roots, and field-grown plant roots, has been reported (Deepa et al., Citation2014; Matsunami et al., Citation2018; Muoki et al., Citation2012). In our previous study on RNA extraction from paddy-grown rice roots, the total RNA amounts decreased as plant growth progressed, and it was impossible to extract an adequate amount of RNA from roots after the panicle formation stage using a commercial RNA isolation kit (Matsunami et al., Citation2018). Therefore, we established an effective RNA extraction method (FRP method) that combines a commercial RNA isolation kit and the same reagents with an easy step of purifying the sample and archived to obtain an adequate amount of high-quality RNA from rice roots with a wide growth period (from tillering to ripening).
In this study, we used soybeans because they are one of the major and most economically important upland crops. Soybeans have a main root system and woody type roots, unlike other major cereal crops. There have been previous studies that conducted RNA extraction from soybean roots using a commercial RNA isolation kit and reagents (Prince et al., Citation2019; Yuan et al., Citation2017, Citation2016), but most were extracted from young seedling roots under pot-culture conditions. As with many other previous studies, the details of root sampling and RNA extraction have not well described in the paper. It has been reported that RNA isolation of wood roots is time-consuming and has a very low RNA yield (Muoki et al., Citation2012; Ouyang et al., Citation2014); therefore, it is unclear whether well-developed field-grown soybean roots will cause RNA extraction difficulty. Furthermore, unlike aquatic plants, when upland crop roots are washed with water during the sampling process, the water status of the rhizosphere changed dramatically and could affect gene expression. However, it is not clear how long washing will affect gene expression. The goal of this study was to examine whether the root-sampling process (time of washing roots with water) affects gene expression analysis, and the difficulty of extracting RNA from field-grown soybean roots exists and whether the FRP method is effective for the RNA extraction from soybean roots.
Materials and methods
Pot experiment
Soybeans (Glycine max (L.) Merr. Nanbushirome) was sown into a pot, one plant per pot. The pot size was φ12.7 cm width and 19.8 cm height. To remove the soil from the roots quickly, river sand was filled in pots to 5 cm from the top. A soybean seed was sown on the sand and then covered with seedling culture soil. The plants were grown in a glasshouse. Water was applied when the surface of the soil was dry.
At 21 DAS, the roots were sampled using the following methods. Roots were washed with tap water for 5 s, 1 min, and 5 min. The sand was easily and immediately removed by submersion in water. After washing, the roots were dried on a paper towel, wrapped with aluminum foil, and immediately frozen in liquid nitrogen. The root samples were ground using a mortar and pestle in liquid nitrogen and stored at −80°C until RNA extraction.
For the extraction of total RNA from the root samples, we used the RNeasy Plant Mini Kit (Qiagen, The Netherlands) alone or in combination with Fruit-mate for RNA Purification (Takara Bio Inc., Japan) and RNAiso Plus (Takara Bio Inc., Japan).
RNeasy Plant Mini Kit method (PMK): RNA was extracted from approximately 100 mg of ground root tissue using the RNeasy PMK according to the manufacturer’s instructions.
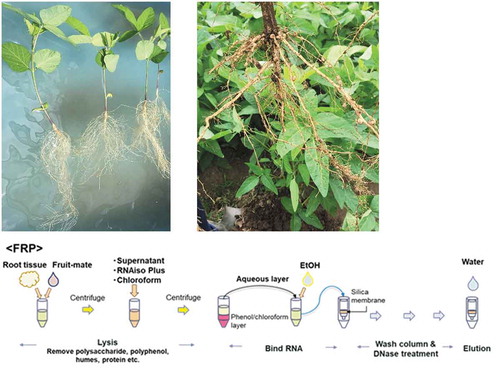
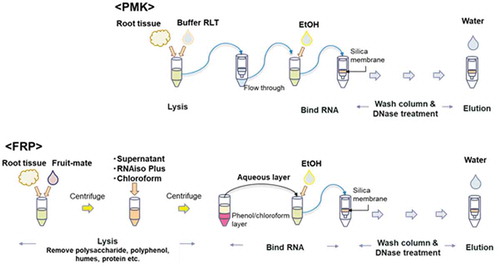
Combination of Fruit-mate for RNA Purification, RNAiso Plus, and RNeasy Plant Mini Kit (FRP): RNA was extracted from approximately 500 mg of ground roots. In this method, samples were pretreated with Fruit-mate for RNA Purification to purify the sample, and then RNA was lysed by the acid guanidinium-phenol-chloroform method using RNAiso Plus. Next, RNA was washed and treated with RNase-Free DNase I (Qiagen, The Netherlands) using the column attached to the RNeasy PMK. Further details of the FRP method are described in Matsunami et al. (Citation2018). A simple flow chart of the PMK and FRP methods is shown in .
Figure 1. Flow chart showing the procedure for the PMK and FRP methods. This chart is a modified version of the chart shown in.Matsunami et al. (Citation2018)
The concentration of RNA was quantified in a spectrophotometer (Nano Drop 1000, Thermo Scientific, Japan). The purity of RNA was estimated using the A260/A280 and A260/A230 ratio. The quality of RNA was assessed by agarose gel electrophoresis.
First-strand cDNA was synthesized from 0.5 μg of RNA using the ReverTra Ace qPCR RT Kit (TOYOBO, Japan), according to the manufacturer’s instructions. Real-time PCR was performed using the CFX Connect Real-Time PCR Detection System (BIO-RAD, Japan). The PCR conditions were as follows: 95°C for 20 s, followed by 40 cycles of 95°C for 3 s, and 60°C for 30 s. The melt-curve reaction was conducted using the methods recommended by the instrument. To confirm that there was no contamination of genomic DNA in the extracted RNA, we conducted real-time PCR using the RNA solutions as templates. We concluded there was no DNA contamination if essentially no amplification was observed in the PCR. Relative expression levels of nine soybeans aquaporins were calculated by the ΔΔCt method using 18s rRNA as a reference. Primer sequences are listed in . The nine aquaporins shown higher level of expression in roots among 45 aquaporins (Ishikawa, Citation2016). To calculate significant differences between wash-treatments, the data were at first transformed by Box-Cox transformation, then Dunnett’s test was conducted by using JMP 14 software.
Table 1. Primer sequences of soybean aquaporins detected in this study
Field experiment
We determined whether the RNA extraction from field-grown soybean roots is possible by the methods (PMK and FRP) described above. Soybeans (Glycine max (L.) Merr. Nanbushirome) was sown by hand in the experimental field at Iwate University on 17 May 2019. The density of sowing was 9.5 plants m−2, with 0.15 m between hills and 0.70 m between rows. Two seeds were sown and seedlings were thinned to one per hill after establishment. Fertilizer was not applied. A pre-emergence herbicide (Basta, BSDF Japan Inc., Tokyo) was applied immediately after sowing according to the manufacturer’s recommendations.
Sample collection was performed at 35 (vegetative stage), 56 (vegetative stage), 76 (flowering stage), and 97 (seed filling) d after sowing (DAS). Flowering started at 61 DAS. Sampling was completed from 10:00–12:00 on sunny days. Roots were collected with a shovel (approximately 30 cm depth), the soil block was removed from the roots by hand, and they were lightly washed with tap water to remove the remaining soil. The main root, except the enlarged part and lateral roots were cut with scissors, patted dry on a paper towel, wrapped with aluminum foil, and immediately frozen in liquid nitrogen. The number of collected plants was 18–24, depending on the root size. The roots were ground and mixed using a mortar and pestle in liquid nitrogen and placed in a 50 mL collection tube. Each tube contained 3–6 plants and was prepared in triplicate from different plants. The samples were stored at −80°C until use for RNA extraction. Total RNA was extracted by PMK and FRP methods and then the RNA concentration and purity were checked as described above.
Results and discussion
RNA extraction from soybean roots
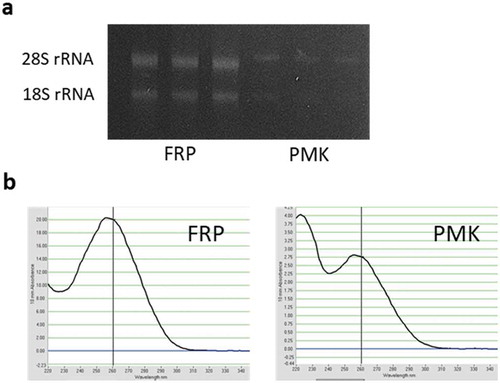
shows the concentration and amount of total RNA collected from the soybean root samples. We empirically observed unstable PCR analysis results when using the RNA sample with a low concentration (below 400 ng/µl); therefore, in our experimental system, adequate quantities of total RNA were at least 400 ng/µl for downstream application. From the pot-grown soybean roots, the extracted RNA concentration by PMK was low (RNA concentration mean was 106 ng/µl), and the ratio of A260/A230 was low (the ratio = 1.2). If the value is lower than 2, then it indicates that contaminants such as carbohydrates, EDTA, guanidine isothiocyanate and phenol that absorb at 230 nm are present in the sample (Sah et al., Citation2014). We could successfully obtain high concentration of total RNA (1381 ng/µl) by using FRP method. Agarose gel electrophoresis of RNA showed two distinct visible bands, indicating the quality of RNA was high ()), although the bands were thin in PMK method. The reason for the lack of extraction of RNA by the PMK method was not elucidated in this study, but one possible explanation was because we used sand for cultivation, which may have caused water stress. Under water-stressed conditions, soybean roots enhance the accumulation of lignin (Yamaguchi et al., Citation2010), which is a major contaminant of RNA extraction. In addition, the high A230 was observed in the RNA sample extracted by PMK ()). The accumulation of soluble sugars was induced by osmotic stresses (Darko et al., Citation2019), thus, this may cause the lower level of purity of RNA. Therefore, accumulation of these substances may have reduced RNA yield and purity. During the process of FRP method, the interfering substances can be removed by pretreatment reagents and centrifugation processes. Thus, the FRP method was shown to be effective in samples with high concentration of interfering substances.
Table 2. The concentration of total RNA extracted from soybean roots and the RNA purity
Figure 2. Quality and purity of total RNA isolated from pot-grown soybean roots extracted by FRP and PMK methods. a Agarose gel electrophoresis of RNA. b RNA spectrum
From the field-grown soybean roots, however, adequate quantities and high purity of total RNA could extract regardless of the extraction method (). By PMK method, the highest concentration was 933 ng/µl at 35 DAS, and the concentration decreased at 56, 76, and 97 DAS (568–627 ng/µl) compared with that at 35 DAS. The FRP method was also effective in extracting RNA; the concentration from the 76 and 97 DAS samples were 1805 and 2160 ng/µl, respectively. In our previous study, the RNA quantities extracted from paddy-grown rice roots dramatically decreased as growth progressed (Matsunami et al., Citation2018); the collected RNA was 404 ng/µl at 34 days after transplanting (DAT) (tillering stage) and 34 ng/µl at 58 DAT (panicle formation stage) using the PMK method. We considered that the low yield of total RNA from field-grown rice roots was caused by a low ratio of active cells in the total root volume. A mature root has lysigenous aerenchyma (programmed cell death), which occupies a large part of the roots (Kawai et al., Citation1998; Suralta & Yamauchi, Citation2008). Furthermore, the architecture of the rice roots is fibrous root system; thus, it is difficult to separate the active part (e.g., apical part of nodal/lateral roots) within a few minutes under field conditions because the time consumption can affect gene expression. When we collected rice roots from the paddies, the root samples were a mixture of aged/young roots and apical/basal parts; thus, the active cell ratio decreased as root growth (and senescence) progressed. In contrast, because soybeans grow under upland conditions and have a main root system, it is easier to collect the roots from wider and deeper soil, and to collect lateral roots which is considered as the active part. We expected that RNA extraction from well-developed soybean roots would be difficult, but in practice, it was possible to extract adequate total RNA from soybean roots with wide growth period.
Root-washing time affects gene expression of aquaporin
The root sampling is an important step in gene expression analysis because the method and time may affect gene expression. To collect the root samples from upland crops, the immersion of roots in water likely affects gene expression because of the root water status changes with washing. Therefore, in a previous study, roots were cleaned by shaking and then frozen in liquid nitrogen (Henry et al., Citation2012). However, to minimize the effect of soil contamination on RNA extraction and downstream applications (Wang et al., Citation2012), obtaining a clean sample is desirable.
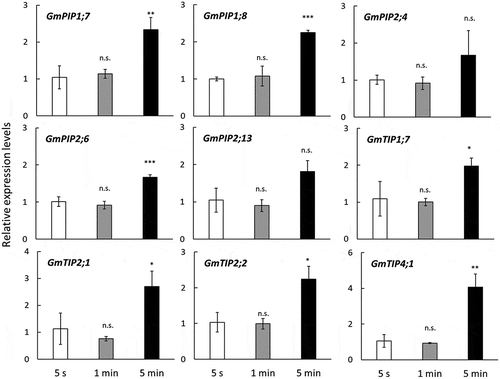
shows the effect of root-washing time on the gene expression levels of aquaporins. Because we considered that 5 s of washing may not affect gene expression, we used the 5 s regime as the control. The expression of aquaporin did not change after 1 min of washing but was affected by washing for 5 min as compared with that of the control (5 s). Although this study analyzed only several aquaporin genes, these responses to moisture condition changes may occur in other genes. Therefore, it is recommended not to take more than 1 min to wash the roots with water. We removed soil from field-grown soybean roots by hand after excavation; thus, washing could conduct within 1 min. However, plants that have fibrous root systems (e.g., monocotyledonous plants) will take more time to remove soil, especially at the basal part. The paddy-grown rice roots at the heading stage took 1.5 min–2.5 min to wash, even if the skillful person conducted the washing. Time consumption during root washing depends on the crop species, growth stage, and soil type; thus, it is important to establish an efficient sampling method for each target to obtain RNA that reflects the correct gene expression profile.
Figure 3. Effect of time for root washing with water on root aquaporin expression levels. Error bars indicate standard deviation (n = 3). The data were transformed by Box-Cox transformation, then conducted the Dunnett’s test. Asterisks indicate significant differences between the 5 s and the 1- or 5-min intervals. A significant effect of washing at the 0.001 (***), 0.01 (**) and 0.05 (*) probability levels. n.s. means not significantly different
In conclusion, this study demonstrated the effectiveness of the FRP method for both field- and pot-cultured soybean roots to extract total RNA. Using pretreatment regents does not significantly increase the cost of extraction but increase extraction success rate. That is, if the RNA is difficult to extract (from mature and old roots, or high interfering substances contaminated sample), the cost and time can be saved by reducing the number of failures. It is difficult to collect the root system and obtain the high quality of RNA sample under field conditions. In this study, we suggested that the washing process may affect gene expression, but there are other points to be noted, such as the time of sampling, time after cutting to freeze sample etc. These technical effects should be considered for field-based gene expression analysis. It is important to reveal gene expression throughout a wide range of growth periods, from not only young seedlings or early vegetative stage plants, to evaluate the function of genes and their age-dependence. Field-based gene expression analysis and transcriptome provide us the deeper understanding of gene function and its practical application for crop production.
Acknowledgments
We thank Dr. Junko Ishikawa-Sakurai (NARO Institute of Crop Science, NARO) for providing information on soybean aquaporin and its primer sequence. We also thank Dr. Katsunori Hatakeyama and Ms. Noriko Yamagishi (Iwate University) for their valuable comments on this study.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Darko, E., Végh, B., Khalil, R., Marček, T., Szalai, G., Pál, M., & Janda, T. (2019). Metabolic responses of wheat seedlings to osmotic stress induced by various osmolytes under iso-osmotic conditions. PLoS ONE, 14(12), e0226151. https://doi.org/10.1371/journal.pone.0226151
- Deepa, K., Sheeja, T. E., Santhi, R., Sasikumar, B., Cyriac, A., Deepesh, P. V., & Prasath, D. (2014). A simple and efficient protocol for isolation of high quality functional RNA from different tissues of turmeric (Curcuma longa L.). Physiology and Molecular Biology of Plants, 20(2), 263–271. https://doi.org/10.1007/s12298-013-0218-y
- Henry, A., Cal, A. J., Batoto, T. C., Torres, R. O., & Serraj, R. (2012). Root attributes affecting water uptake of rice (Oryza sativa) under drought. Journal of Experimental Botany, 63(13), 4751–4763. https://doi.org/10.1093/jxb/ers150
- Ishikawa, J. (2016) Role of soybean aquaporins in inhibition of water uptake under flooding. KAKEN Final Research Report. *in japanese with English abstract. https://kaken.nii.ac.jp/ja/file/KAKENHI-PROJECT-25450031/25450031seika.pdf
- Kawai, M., Smamarajeewa, P. K., Barrero, R. A., Nishiguchi, M., & Uchiyama, H. (1998). Cellular dissection of the degradation pattern of cortical cell death during aerenchyma formation of rice roots. Planta, 204(3), 277–287. https://doi.org/10.1007/s004250050257
- Matsunami, M., Hayashi, H., Tominaga, Y., Nagamura, Y., Murai-Hatano, M., & Ishikawa-Sakurai, J. (2018). Effective methods for practical application of gene expression analysis in field-grown rice roots. Plant and Soil, 433(1–2), 173–187. https://doi.org/10.1007/s11104-018-3834-z
- Muoki, R.C., Paul, A., Kumari, A., Singh, K. & Kumar, S. An improved protocol for the isolation of RNA from roots of tea (Camellia sinensis (L.) O. Kuntze). Mol Biotechnol. 2012 Sep; 52(1), 82–88. doi: 10.1007/s12033-011-9476-5. PMID: 22144070
- Nagano, A. J., Sato, Y., Mihara, M., Antonio, B. A., Motoyama, R., Itoh, H., Nagamura, Y. & Izawa, T. (2012) Deciphering and prediction of transcriptome dynamics under fluctuating field conditions. Cell 141(6), 1358–1369. https://doi.org/10.1016/j.cell.2012.10.048
- Ouyang, K., Li, J., Huang, H., Que, Q., Li, P., & Chen, X. (2014). A simple method for RNA isolation from various tissues of the tree Neolamarckia cadamba. Biotechnology & Biotechnological Equipment, 28(6), 1008–1013. https://doi.org/10.1080/13102818.2014.981086
- Prince, S. J., Valliyodan, B., Ye, H., Yang, M., Tai, S., Hu, W., Murphy, M., Durnell, L. A., Song, L., Joshi, T., Liu, Y., Van de Velde, J., Vandepoele, K., Grover Shannon, J., & Nguyen, H. T. (2019). Understanding genetic control of root system architecture in soybean: Insights into the genetic basis of lateral root number. Plant, Cell & Environment, 42(1), 212–229. https://doi.org/10.1111/pce.13333
- Sah, S. K., Kaur, G., & Kaur, A. (2014). Rapid and reliable method of high-quality RNA extraction from diverse plants. American Journal of Plant Sciences, 5(21), 3129–3139. https://doi.org/10.4236/ajps.2014.521329
- Suralta, R. R., & Yamauchi, A. (2008). Root growth, aerenchyma development, and oxygen transport in rice genotypes subjected to drought and waterlogging. Environmental and Experimental Botany, 64(1), 75–82. https://doi.org/10.1016/j.envexpbot.2008.01.004
- Wang, Y., Hayatsu, M., & Fujii, T. (2012). Extraction of bacterial RNA from soil: Challenges and solutions. Microbes Environment, 27(2), 111–121. https://doi.org/10.1264/jsme2.ME11304
- Yamaguchi, M., Valliyodan, B., Zhang, J., Lenoble, M. E., Yu, O., Rogers, E. E., Nguyen, H. T., & Sharp, R. E. (2010). Regulation of growth response to water stress in the soybean primary root. I. Proteomic analysis reveals region‐specific regulation of phenylpropanoid metabolism and control of free iron in the elongation zone. Plant, Cell & Environment, 33(2), 223–243. https://doi.org/10.1111/j.1365-3040.2009.02073x
- Yuan, S., Li, R., Chen, H., Zhang, C., Chen ,Li., Hao, Q., Chen, S., Shan, Z., Yang, Z., Zhang, X., Qiu, D., Zhou, X. (2017). RNA-Seq analysis of nodule development at five different developmental stages of soybean (Glycine max) inoculated with Bradyrhizobium japonicum strain 113-2. Scientific Reports, 7, 42248. https://doi.org/10.1038/srep42248
- Yuan, S., Li, R., Chen, S., Chen, H., Zhang, C., Chen, L., Hao, Q., Shan, Z., Yang, Z., Qiu, D., Zhang, X., & Zhou, X. (2016). RNA-Seq analysis of differential gene expression responding to different rhizobium strains in soybean (Glycine max) roots. Frontiers in Plant Science, 7, 721. https://doi.org/10.3389/fpls.2016.00721
- Zhang, D. Y., Ali, Z., Wang, C. B., Xu, L., Yi, J. X., Xu, Z. L., Liu, X. Q., He, X. L., Huang, Y. H., Khan, I. A., Trethowan, R. M., & Ma, H. X. (2013). Genome-wide sequence characterization and expression analysis of major intrinsic proteins in soybean (Glycine max L.). PloS one, 8(2), e56312. https://doi.org/10.1371/journal.pone.0056312