ABSTRACT
Objectives
The study explores the protective role of the peripheral serum of limb remote ischemic postconditioning (LRIP) in reducing the reactive oxygen species (ROS) levels and neutrophil activation, which are responsible for the deleterious reperfusion injury.
Methods
LRIP was induced in Sprague–Dawley rats by three cycles of 5 min occlusion /5 min reperfusion on the left hind limb. The blood samples were collected before LRIP or 0 and 1 h after LRIP (named SerumSham, SerumLRIP0, SerumLRIP1, respectively). The effects of LRIP serum on ROS level and neutrophils activation were determined. The expression of MyD88-TRAF6-MAPKs and PI3K/AKT pathways in neutrophils were examined.
Results
When compared with SerumSham, SerumLRIP0 and SerumLRIP1 significantly reduced the ROS released from neutrophils activated by fMLP. Meanwhile, the mRNA expression levels of NADPH oxidase subunit p22phox and multiple ROS-producing related key proteins, such as NADPH oxidase subunit p47phox ser 304, ser 345. MyD88, p-ERK, p-JNK and p-P38 expression of neutrophils were downregulated by SerumLRIP0 and SerumLRIP1. SerumLRIP1 also downregulated p47phox mRNA expression and tumor necrosis factor receptor-associated factor 6 (TRAF6) protein expression.
Conclusion
LRIP serum protects against ROS level and neutrophils activation involving the MyD88-TRAF6-MAPKs. This finding provides new insight into the understanding of LRIP mechanisms.
GRAPHICAL ABSTRACT
Introduction
Limb remote ischemic postconditioning (LRIP) refers to the repeated limb ischemia after a serious ischemic brain or myocardial injury. LRIP protects against cerebral or myocardial ischemia/reperfusion (I/R) injury [Citation1,Citation2], eventually promoting the recovery in ischemic disease patients. Thus, LRIP has exhibited a promising therapeutic potential. I/R injury is a complicated pathophysiological process that occurs through various pathways including oxidative stress and inflammation. However, the underlying mechanisms remain to be fully elucidated. Recent studies support that the reactive oxygen species (ROS) and proteases released from the migrated neutrophils play a major role in acute reperfusion injury following ischemic stroke [Citation3].
Our previous study revealed that the limb operated with LRIP can ‘attract’ the activated neutrophils, thereby reducing the neutrophils migration to the cerebral ischemic area [Citation3]. Meanwhile, LRIP in rats inhibits the NADPH oxidase activation in neutrophils, a key enzyme which is dedicated to ROS production [Citation4], through TLR4-MyD88-TRAF6 signaling pathway. A recent report presented that the serum extracted from humans who were subjected to remote ischemic preconditioning (RIPC) has a protective effect on human endothelial and intestinal cells against hypoxia-induced damage [Citation5,Citation6]. However, whether the serum of LRIP mediates the protective effect through inhibiting neutrophils activation or migration.
We hypothesized that the LRIP serum from rats could directly reduce the ROS release from neutrophils through inhibiting NADPH oxidase. The associated pathways were examined. The finding contributes to the elucidation of the LRIP protective mechanism.
Materials and methods
Animals
Adult male Sprague–Dawley rats (Qinglongshan, Nanjing, China) weighing 250–280 g were used in this study. All animal experiment protocols were approved by the Animal Ethics Committee of China Pharmaceutical University (No. 81503284) and were conducted according to the guidelines of the Institutional Animal Care and Use Committee at China Pharmaceutical University. Animals were housed in a temperature and light-controlled room (23 ± 1°C; 12 h light/12 h dark cycle) and allowed to freely access to food and water.
Reagents
Hanks’ Balanced Salt Solution (HBSS), Trizol (R0016) and SB203580 (SB, S1863) were obtained from Beyotime Biotechnology Co Ltd. RPMI 1640 and Fetal bovine serum (FBS, 26140-079) were obtained from Wisent Biotechnology Co., Ltd. Formyl-methionyl-leucyl-phenylalanine (fMLP, C-1711) and Apocynin (APO, HY-N0088) were obtained from Sigma-Aldrich and MedChemExpress.
Collection and preparation of LRIP serum of rat
The rats were randomly divided into SerumSham (SSham), SerumLRIP0h (SLRIP0) and SerumLRIP1h (SLRIP1) groups (n = 5). SLRIP0 and SLRIP1 rats were anesthetized with 10% chloral hydrate and placed on the surgical plate. The femoral artery of the left hindlimb of rats was clamped with a modified blood pressure cuff for 5 min (ischemia) and blood flow was restored for 5 min (reperfusion), which was regarded as a cycle and repeated three cycles [Citation3]. Interruption of the blood supply to the hindlimb was confirmed by cyanosis of the skin on the limb. The blood was collected from the abdominal aorta of rats immediately (SLRIP0) or at 1 h after the completion of LRIP (SLRIP1) or without LRIP operation (SSham) [Citation6]. After clotted for 10 min at room temperature, all of the blood samples were then centrifuged at 5000 rpm for 10 min. The serum was collected and stored at −80°C for further use.
Isolation and culture of rat bone marrow neutrophils
The neutrophils were isolated from rat bone marrow and purified following the manufacturer’s instructions of Rat Bone Marrow Isolation Kit (TBD2013NR, Tianjin Haoyang Biological Manufacture CO., Ltd., Tianjin, China). Rats were sacrificed by cervical dislocation under anesthesia. The femur and tibia of rat were isolated and the soft tissue attachment were carefully removed. F solution in Rat Bone Marrow Isolation Kit was forced through the bone with a 10 mL syringe. After dispersing cell clumps, the cell suspension was centrifuged (500 g, 10 min, room temperature) and resuspended in HBSS (Ca2+, Mg2+) solution. Cells were then overlay two-layer solution A gradient of 100% and 80% solution A (diluted in diluent with 100% solution A), and centrifuged (750 g, 25 min, room temperature). Neutrophils from the upper phases were harvested and the remaining red cells were eliminated by red blood cell lysis. After washed twice with cell detergent, the purity of neutrophils was identified by using Wright-Gemssa staining (purity > 90%). The isolated neutrophils were adjusted to 2.5×106/mL with RPMI 1640 complete medium, then subjected to the following treatments and measurements.
Detection of neutrophil viability
To detect the neutrophil viability, cells were separated into 2 groups. In group 1, cells were cultured with RPMI 1640 containing 10% FBS [Citation7]. In group 2, cells were cultured with RPMI 1640 containing 10% SSham at 37 °C under 95% O2, 5% CO2. The neutrophil viability was detected at 0, 3, 6, 12, 24 and 48 h by using the Cell Titer-Lumi™ Cell Viability Assay Kit (C0068S, Beyotime Biotechnology Co., Ltd., Shanghai, China) according to the manufacturer’s instructions.
Release of ROS in neutrophils
To measure the release of ROS and the mRNA/protein levels of associated markers, the neutrophils were randomly distributed into 6 groups: SSham group, SSham + ROS inducer fMLP group, SLRIP0 group, SLRIP1 group, SSham+ p38 MAPK inhibitor SB group, SSham+ NADPH oxidase inhibitor APO group [Citation8,Citation9]. SLRIP0 and SLRIP1 group cultured with RPMI 1640 containing 10% SLRIP0 or 10% SLRIP1, respectively, the other four groups cultured with RPMI 1640 containing 10% SSham. 200 μL cell suspension was inoculated into the 96-well culture plate coated with 0.1% gelatin, then cultured at 37 °C under 95% O2 and 5% CO2. 24 h later, p38 MAPK inhibitor SB (10 μM, DMSO) [Citation10] and NADPH oxidase inhibitor APO (100 μM, DMSO) [Citation11] were added into SB and APO groups, respectively, then SB group was cultured for 30 min and APO group for 15 min, followed by stimulation with fMLP (10 μM, DMSO) [Citation12] for 10 min. After carefully transferred into 96-well plate and gently washed with PBS, the culture medium was added with 100 μL of 2, 7 dichlorofluorescein diacetate (DCFH-DA) diluted (1:1000) to PBS, then incubated at 37°C for 25 min. Fluorescence value was measured at the excitation at 488 nm and emission at 525 nm with an automated microplate reader (Varioskan Flash, ThermoFisher Scientific, Waltham, MA, USA).
qPCR
Total RNA of neutrophils was extracted by Trizol similar as previously described [Citation3]. Then 1 μL cDNA product and 2 μL primer were added into qPCR super mixture with a final volume of 10 μL [Citation3]. qPCR was performed with 1 cycle for 30 s at 94°C, followed by 5 s at 94°C, 40 cycles (1 cycle for 30 s) at 60°C using a Real-time quantitative PCR instrument (Bio-Rad, USA). The following primer sequences were used (5′ to 3′): p47phox (AC008033.9), forward, ATTTTCTTGGTGATTGATG, reverse, GAGGGATGTTACTTACTGG; p22phox (AC008080.1) forward, CAGCCACCCGAGATTGAGCA, reverse, AGGCACGGACAGCAGTAA; GAPDH (LC627775.1), forward, TCCTGCACCACCAACTGCTTAG, reverse, AGTGGCAGTGATGGCATGGACT.
Western blot
Western blot was performed as described previously [Citation3]. The total protein was extracted with cold RIPA lysis buffer from neutrophils and protein concentration was measured using BCA Protein Assay Kit (P0012S, Beyotime Biotechnology Co., Ltd., Shanghai, China), then 40 µg protein from each group were used for protein gel electrophoresis. After the electrophoresis, the protein band was transferred onto the PVDF membrane and blocked with 3% BSA. Followed by incubation with the first antibody overnight at 4°C, (antibodies as below, p47phox (sc-17845, Santa Cruz Biotechnology, Texas, USA), p-p47phox Ser 345 (SAB4301368, Sigma-Aldrich, St. Louis, MO, USA), p-p47phox Ser 304 (P14598, Bioworld, Beijing, China), TRAF6 (ab33915, Abcam, Cambridge, MA, USA), MyD88 (ab133739, Abcam, Cambridge, MA, USA), p38 MAPK (8690S, CST, Danvers, Massachusetts, USA), p-p38 MAPK (4511S, CST, Danvers, Massachusetts, USA), PI3K (17366S, CST, Danvers, Massachusetts, USA), AKT (4685S, CST, Danvers, Massachusetts, USA), p-AKT (4685S, CST, Danvers, Massachusetts, USA), ERK (4695S, CST, Danvers, Massachusetts, USA), p-ERK (4370S, CST, Danvers, Massachusetts, USA), JNK (2305S, CST, Danvers, Massachusetts, USA) and p-JNK (9255S, CST, Danvers, Massachusetts, USA)), the membranes were washed and incubated with the horseradish labeled second antibody for 2 h at room temperature. The protein band was analyzed with the gel-imaging system (Bio-Rad, Hercules, CA, USA). The values were calculated after normalization to the amount of GAPDH.
Statistical analysis
All data was statistically analyzed by using GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA, USA). Results were expressed as mean ± SEM. Student’s t-test was used for the comparison between the two groups. One-way ANOVA analysis was used for the comparison of three or more groups, followed by using Dunnett’s test, P < .05 was considered to be statistically significant.
Results
LRIP serum collection and neutrophil viability determination
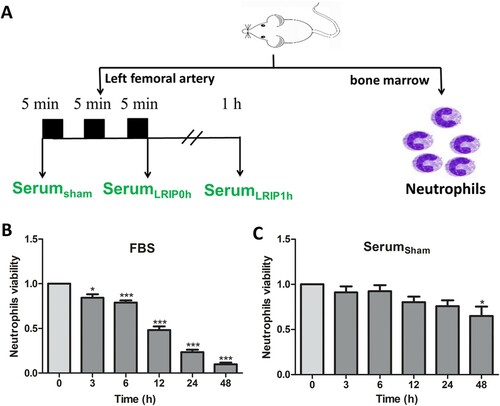
Blood was collected from the abdominal aorta and SSham, SLRIP0 and SLRIP1 were obtained ((A)). Bone marrow neutrophils were separated and the quality was determined by Wright-Giemsa staining assay (purity > 90%). The neutrophil viability was evaluated at 0, 3, 6, 12, 24 and 48 h to compare the effect of rat serum and FBS. Results showed that the viability of neutrophils cultured with FBS significantly decreased at 3 h compared with viability at 0 h ((B)) (P < .05). It is interesting to find that neutrophils cultured with rat serum still remained at 90% viability (0 h viability as 100%) and remained at higher viability even at 48 h ((C)). Based on these results, we selected 24 h as the time point for testing the effects of LRIP serums on neutrophils.
Figure 1. Combine LRIP serum collecting and neutrophil viability cultured in vitro. (A) Blood was collected before LRIP operation, or 0, 1 h after LRIP operation, serum was named SSham, SLRIP0, SLRIP1 separately. The viability of bone marrow neutrophils was assessed at 0, 3, 6, 12, 24, 48 h under different serum conditions:(B) Cultured with FBS; (C) Cultured with SSham. Data are expressed as mean ± SEM, n = 4–5, ***P < .001 vs. 0 h, *P < .05 vs. 0 h.
LRIP serum inhibits fMLP-induced neutrophil ROS release
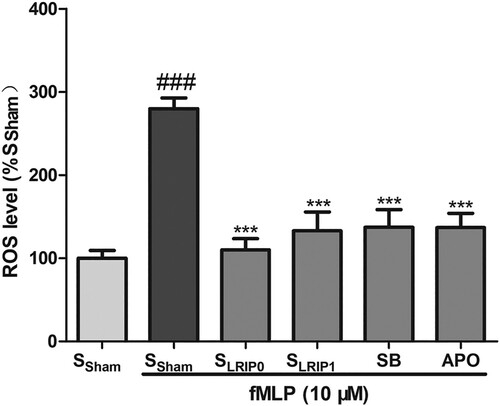
Compared with SSham group, SSham + fMLP group significantly increased the excessive of ROS (P < .001). SLRIP0, SLRIP1, SB203580 and APO all could significantly inhibit ROS production ().
Figure 2. The inhibiting effects of LRIP serum on intracellular superoxide generation from neutrophils. Neutrophils were cultured with RPMI 1640 containing SSham, SLRIP0 or SLRIP1 serum for 24 h, followed by incubating with DMSO, Apocynin (100 μM) for 15 min or incubated with SB203580 (10 μM) for 30 min, then activated with fMLP (10 μM) for 10 min. Data are expressed as mean ± SEM, n = 5, ###P < .001 vs. SSham *** P < .0001 vs. SSham + F.
LRIP serum inhibits the activation of neutrophil NADPH oxidase induced by fMLP
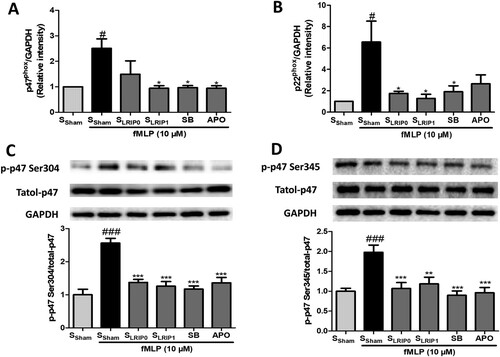
The mRNA expression of p47phox and p22phox, two key regulators of NADPH oxidase, were significantly upregulated in the SSham + fMLP group (P < .05). Similar to the SSham group, the phosphorylation of p47phox Ser 304 and p47phox Ser 345 were also significantly upregulated (), p47phox mRNA of SLRIP1 + fMLP, SB and APO groups, as well as p22phox mRNA of SLRIP0, SLRIP1 and SB groups were significantly downregulated (P < .05). SLRIP0, SLRIP1, SB and APO all could significantly inhibited the phosphorylation of p47phox Ser 304 and Ser 345. These results indicated that LRIP serum can inhibit the activation of neutrophil NADPH oxidase induced by fMLP.
Figure 3. The inhibiting effects of LRIP serum on activation of neutrophil NADPH oxidase. qPCR results showed the mRNA expression of p47phox (A) and (B) p22phox, GAPDH was used as the internal control, (n = 3); (C–D) Western bolt analysis of p47phox phosphorylation at Ser 304 and Ser 345 of neutrophils (n = 5). Data are expressed as mean ± SEM, ###P < .001 vs. SSham, #P < .05 vs. SSham, ***P < .001 vs. SSham + F, **P < .01 vs. SSham + F, *P < .05 vs. SSham + F.
LRIP serum inhibits the activation of neutrophil NADPH oxidase induced by fMLP
To determine whether LRIP serum inhibits the activation of neutrophil NADPH oxidase induced by fMLP, neutrophils were treated with SSham, SLRIP0, SLRIP1, SB and APO before being activated by fMLP. Compared with SSham group, the mRNA expression of p47phox and p22phox, as well as the phosphorylation of p47phox Ser 304 and p47phox Ser 345 was significantly increased in the SSham + fMLP group (P < .05, ). SLRIP1, SB and APO all could downregulate the expression of p47phox mRNA (P < .05). SLRIP0, SLRIP1, SB and APO all could significantly inhibit the phosphorylation of p47phox Ser 304 and Ser 345, SLRIP0, SLRIP1, SB also could downregulate the expression of p22phox mRNA (P < .05). These results indicated that LRIP serum could inhibit the activation of neutrophil NADPH oxidase induced by fMLP.
LRIP serum inhibits the activation neutrophil induced by fMLP via MyD88-TRAF6-MAPKs signaling pathway
Compared with SSham group, the protein expression of MyD88, TRAF6, p-ERK, p-JNK and p-p38 MAPK of SSham + fMLP groups were significantly upregulated. SLRIP0, SLRIP1, SB and APO all could downregulated the protein expression of MyD88, TRAF6, p-ERK, p-JNK and p-p38 MAPK. These results proved that the LRIP serum could inhibit the activation of neutrophil NADPH oxidase induced by fMLP via the MyD88-TRAF6-MAPKs signaling pathway ().
Figure 4. The inhibiting effects of LRIP serum on fMLP-induced neutrophil activation via MyD88/TRAF6/p38MAPK pathway. Representative images of Western bolt assessments and quantitative analysis of the ratio of MyD88 (A); TRAF6 (B); p-ERK (C); p-JNK (D); and p-p38 MAPK (E). Data are expressed as mean ± SEM, n = 5, ###P < .001 vs. SSham, #P < .05 vs. SSham, ***P < .001 vs. SSham + F, **P < .01 vs. SSham + F, *P < .05 vs. SSham + F.
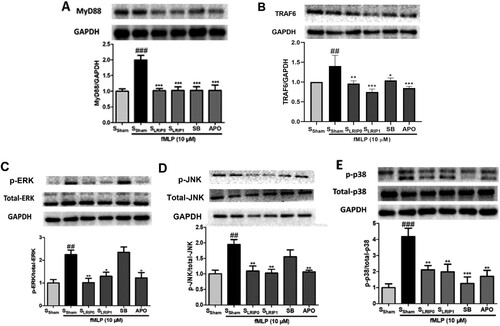
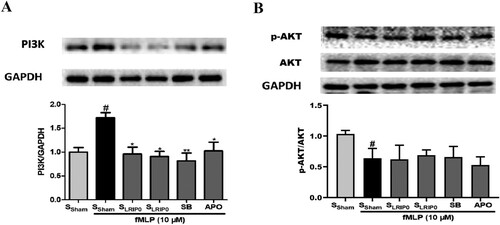
LRIP serum inhibits the activation of neutrophil induced by fMLP independently of PI3 K/AKT signaling pathway
Continuously, to determine whether PI3K/Akt pathway is involved. As shown in , compared with the SSham group, the expression of PI3K of the SSham + fMLP group was significantly upregulated but the p-Akt expression levels were downregulated by fMLP. SLRIP0, SLRIP1, SB and APO all could downregulate the expression of PI3K. These results revealed that LRIP serum inhibits the activation of neutrophil induced by fMLP, the PI3K may play a role in it but independently of p-AKT.
Figure 5. The inhibiting effects of LRIP serum independently of PI3K/Akt in fMLP-induced neutrophil. Representative images of Western bolt assessments and quantitative analysis of the ratio of: PI3K (A); p-AKT/Akt (B). Data are expressed as mean ± SEM, n = 4, #P < .05 vs. SSham, **P < .01 vs. SSham + F, *P < .05 vs. SSham + F.
Discussion
In this study, we wanted to test the effect and mechanism of LRIP serum on a neutrophil respiratory burst model in vitro. The lifespan of neutrophils in culture with FBS or rat serum was tested. Recent studies have confirmed that the lifespan of neutrophils in the blood circulation of mice is 18 h [Citation13], and in humans, neutrophils can survive in the blood for 5.4 days [Citation14]. Previous research has proved that there are a great amount of functionally competent neutrophils in mice bone marrow, the half-life of neutrophils from bone marrow is longer than the neutrophils from blood [Citation15]. This study showed that the activity of neutrophils cultured with FBS decreased significantly after 3 h, this is similar to previous studies [Citation15]. It is very interesting to find that neutrophils cultured with SSham remained more than 90% viability after a 24 h culture (). This proved that there should be some nutritional factors in SSham, which satisfied the rat neutrophil survival needs. Based on this result, we chose neutrophils extracted from the bone marrow and the rat serum as the cell culture media, 24 h as the culture time in this study. In addition, SLRIP0, SLRIP1, SB and APO were all found to inhibit the ROS production from neutrophils activated by fMLP (). This result proved the inhibiting effect of SLRIP0, SLRIP1 on the ROS release from neutrophils under the stimulated I/R condition and also proved LRIP can exert protective effects through the serum route. Futhermore, results of qPCR and western blot showed that SLRIP1 could downregulate the mRNA expression of p47phox. Both SLRIP0 and SLRIP1 could downregulate the mRNA and protein expression of p22phox, p47phox ser 304 and p47phox ser 345 ().
ROS was thought to be the major cause of I/R injury [Citation16–18] and the activated neutrophil NADPH oxidase was thought to be the main source of ROS [Citation19–21]. The reperfusion injury would occur [Citation22,Citation23] after the thrombolytic therapy was used on ischemic disease patients to restore blood flow into the ischemic regions [Citation24,Citation25]. LRIP is effective in alleviating I/R injury of brain, heart, kidney and other organs [Citation3,Citation26,Citation27]. The related mechanisms include reversing the eNOS uncoupling [Citation28], upregulating the expression of Nrf2 along with heme oxygenase-1 (HO-1) [Citation29], quinone oxidoreductase 1 (NQO-1) [Citation30], inhibiting the inflammation [Citation31–33]. Our previous work has proved that LRIP can inhibit the activation of neutrophil NADPH oxidase through the TLR4-MyD88-TRAF6 pathway, working to reduce the cerebral I/R injury [Citation3]. But till now, no researches have been done to clarify the effect and mechanism of LRIP serum in the activation of neutrophil NADPH oxidase in vitro. In this study, we wanted to check the protective effect of LRIP serum and clarify the related mechanisms.
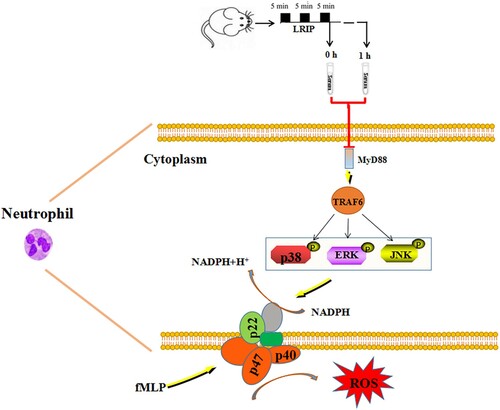
The activation of neutrophil NADPH oxidase induced by fMLP will produce a great deal of ROS [Citation34], which can simulate the respiratory burst of neutrophils under I/R injury. In our study, 10 μM fMLP was used to stimulate neutrophils [Citation35]. The catalytic core of NADPH oxidase in neutrophils is gp91phox (Nox2). Activation of Nox2 needs the active p47phox (the adaptor protein) to bind onto p22phox [Citation36] and p67phox onto Nox2. The function of p47phox is governed by serine phosphorylation, which occurs during activation of the NADPH oxidase. Phosphorylation of Ser-304 [Citation37] and Ser-345 [Citation38] on p47phox is a necessary event in the priming of neutrophils. Results in this study proved fMLP could upregulate the mRNA expression of p22phox, p47phox in neutrophils and upregulate the phosphorylation of ERK, JNK, P38, p47phox ser 304, ser 345, also upregulate the expression of MyD88, TRAF6 in neutrophils. These proved the activation of neutrophil NADPH oxidase simulated by fMLP was based on the MyD88/TRAF6/p38 MAPK pathway (). The results also showed that this kind of activation can be inhibited by SLRIP0 and SLRIP1. This demonstrates that SLRIP0 and SLRIP1 inhibited the production of ROS from activated neutrophils by inhibiting the activation of neutrophil NADPH oxidase.
Figure 6. Serum of LRIP inhibits fMLP-triggered activation and ROS releasing of rat neutrophils through MyD88/TRAF6/p38 MAPK pathway.
This study still has some limitations. For example, the kind of serum factor/factors which mediated the protective effect of the LRIP serum and how PI3K regulated the protective effect of LRIP serum, both remain to be identified.
Conclusion
In conclusion, the rat serum is more adept to keeping viability of the bone marrow neutrophils. LRIP operation can induce some serum factors to inhibit the activation of neutrophil NADPH oxidase via the MyD88/TRAF6/p38 MAPK pathway, which then alleviate the I/R injury. This study might be meaningful for clarifying the protection mechanisms of LRIP and allowing for further research and development of biological drugs related to I/R injury.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Guo H, Zhao L, Wang B, et al. Remote limb ischemic postconditioning protects against cerebral ischemia-reperfusion injury by activating AMPK-dependent autophagy. Brain Res Bull. 2018;139:105–113. doi:https://doi.org/10.1016/j.brainresbull.2018.02.013.
- Liang D, He XB, Wang Z, et al. Remote limb ischemic postconditioning promotes motor function recovery in a rat model of ischemic stroke via the up-regulation of endogenous tissue kallikrein. CNS Neurosci Ther. 2018;24:519–527. doi:https://doi.org/10.1111/cns.12813.
- Chen G, Ye X, Zhang J, et al. Limb remote ischemic postconditioning reduces ischemia-reperfusion injury by inhibiting NADPH oxidase activation and MyD88-TRAF6-P38MAP-kinase pathway of neutrophils. Int J Mol Sci. 2016;17; doi:https://doi.org/10.3390/ijms17121971.
- Magnani F, Nenci S, Millana Fananas E, et al. Crystal structures and atomic model of NADPH oxidase. Proc Natl Acad Sci U S A. 2017;114:6764–6769. doi:https://doi.org/10.1073/pnas.1702293114.
- Zitta K, Meybohm P, Bein B, et al. Serum from patients undergoing remote ischemic preconditioning protects cultured human intestinal cells from hypoxia-induced damage: involvement of matrixmetalloproteinase-2 and -9. Mol Med. 2012;18:29–37. doi:https://doi.org/10.2119/molmed.2011.00278.
- Weber NC, Riedemann I, Smit KF, et al. Plasma from human volunteers subjected to remote ischemic preconditioning protects human endothelial cells from hypoxia-induced cell damage. Basic Res Cardiol. 2015;110:17, doi:https://doi.org/10.1007/s00395-015-0474-9.
- Xu T, Cai Y, Zhong X, et al. β-Galactosidase instructed supramolecular hydrogelation for selective identification and removal of senescent cells. Chem Commun (Camb). 2019;55:7175–7178. doi:https://doi.org/10.1039/c9cc03056e.
- Ye W, Zhong Z, Zhu S, et al. Advanced oxidation protein products induce catabolic effect through oxidant-dependent activation of NF-κ B pathway in human chondrocyte. Int Immunopharmacol. 2016;39:149–157. doi:https://doi.org/10.1016/j.intimp.2016.07.018.
- Vorobjeva N, Prikhodko A, Galkin I, et al. Mitochondrial reactive oxygen species are involved in chemoattractant-induced oxidative burst and degranulation of human neutrophils in vitro. Eur J Cell Biol. 2017;96:254–265. doi:https://doi.org/10.1016/j.ejcb.2017.03.003.
- Xia L, Zhou H, Wang T, et al. Activation of mTOR is involved in anti-β2GPI/β2GPI-induced expression of tissue factor and IL-8 in monocytes. Thromb Res. 2017;157:103–110. doi:https://doi.org/10.1016/j.thromres.2017.05.023.
- Wolf ST, Jablonski NG, Ferguson SB, et al. Four weeks of vitamin D supplementation improves nitric oxide-mediated microvascular function in college-aged African Americans. Am J Physiol Heart Circ Physiol. 2020;319:H906–906H914. doi:https://doi.org/10.1152/ajpheart.00631.2020.
- Shi B, Song D, Xue H, et al. Abnormal expression of the peptide transporter PepT1 in the colon of massive bowel resection rat: a potential route for colonic mucosa damage by transport of fMLP. Dig Dis Sci. 2006;51:2087–2093. doi:https://doi.org/10.1007/s10620-005-9067-z.
- Pillay J, den Braber I, Vrisekoop N, et al. In vivo labeling with 2H2O reveals a human neutrophil lifespan of 5.4 days. Blood. 2010;116:625–627. doi:https://doi.org/10.1182/blood-2010-01-259028.
- Day RB, Link DC. Regulation of neutrophil trafficking from the bone marrow. Cell Mol Life Sci. 2012;69:1415–1423. doi:https://doi.org/10.1007/s00018-011-0870-8.
- Boxio R, Bossenmeyer-Pourié C, Steinckwich N, et al. Mouse bone marrow contains large numbers of functionally competent neutrophils. J Leukoc Biol. 2004;75:604–611. doi:https://doi.org/10.1189/jlb.0703340.
- Jiang M, Ni J, Cao Y, et al. Astragaloside IV attenuates Myocardial ischemia-reperfusion injury from oxidative stress by regulating succinate, lysophospholipid metabolism, and ROS scavenging system. Oxid Med Cell Longev. 2019;2019:9137654), doi:https://doi.org/10.1155/2019/9137654.
- Jun JH, Shim JK, Oh JE, et al. Protective effect of ethyl pyruvate against Myocardial Ischemia reperfusion injury through regulations of ROS-related NLRP3 inflammasome activation. Oxid Med Cell Longev. 2019;2019:4264580), doi:https://doi.org/10.1155/2019/4264580.
- Wallert M, Ziegler M, Wang X, et al. α-Tocopherol preserves cardiac function by reducing oxidative stress and inflammation in ischemia/reperfusion injury. Redox Biol. 2019;26:101292. doi:https://doi.org/10.1016/j.redox.2019.101292.
- Yang Z, Sharma AK, Marshall M, et al. NADPH oxidase in bone marrow-derived cells mediates pulmonary ischemia-reperfusion injury. Am J Respir Cell Mol Biol. 2009;40:375–381. doi:https://doi.org/10.1165/rcmb.2008-0300OC.
- Sun L, Wu Q, Nie Y, et al. A role for MK2 in enhancing neutrophil-derived ROS production and aggravating liver ischemia/reperfusion injury. Front Immunol. 2018;9:2610), doi:https://doi.org/10.3389/fimmu.2018.02610.
- Yao J, Zheng J, Cai J, et al. Extracellular vesicles derived from human umbilical cord mesenchymal stem cells alleviate rat hepatic ischemia-reperfusion injury by suppressing oxidative stress and neutrophil inflammatory response. FASEB J. 2019;33:1695–1710. doi:https://doi.org/10.1096/fj.201800131RR.
- Wang G, Weng YC, Han X, et al. Lipocalin-2 released in response to cerebral ischaemia mediates reperfusion injury in mice. J Cell Mol Med. 2015;19:1637–1645. doi:https://doi.org/10.1111/jcmm.12538.
- Zarisfi M, Allahtavakoli F, Hassanipour M, et al. Transient brain hypothermia reduces the reperfusion injury of delayed tissue plasminogen activator and extends its therapeutic time window in a focal embolic stroke model. Brain Res Bull. 2017;134:85–90. doi:https://doi.org/10.1016/j.brainresbull.2017.07.007.
- Chen QF, Liu YY, Pan CS, et al. Angioedema and hemorrhage after 4.5-hour tPA (tissue-type plasminogen activator) Thrombolysis ameliorated by T541 via restoring brain microvascular integrity. Stroke. 2018;49:2211–2219. doi:https://doi.org/10.1161/STROKEAHA.118.021754.
- Lerario MP, Grotta JC, Merkler AE, et al. Association between intravenous Thrombolysis and anaphylaxis Among medicare beneficiaries With acute ischemic stroke. Stroke. 2019;50:3283–3285. STROKEAHA119026861. doi:https://doi.org/10.1161/STROKEAHA.119.026861.
- Gao Y, Song J, Chen H, et al. TRPV1 activation is involved in the cardioprotection of remote limb ischemic postconditioning in ischemia-reperfusion injury rats. Biochem Biophys Res Commun. 2015;463:1034–1039. doi:https://doi.org/10.1016/j.bbrc.2015.06.054.
- Zhou CC, Yao WT, Ge YZ, et al. Remote ischemic conditioning for the prevention of contrast-induced acute kidney injury in patients undergoing intravascular contrast administration: a meta-analysis and trial sequential analysis of 16 randomized controlled trials. Oncotarget. 2017;8:79323–79336. doi:https://doi.org/10.18632/oncotarget.18106.
- Chen G, Yang J, Lu G, et al. Limb remote ischemic post-conditioning reduces brain reperfusion injury by reversing eNOS uncoupling. Indian J Exp Biol. 2014;52:597–605.
- Gao S, Zhan L, Yang Z, et al. Remote limb ischaemic postconditioning protects against Myocardial ischaemia/reperfusion injury in mice: activation of JAK/STAT3-mediated Nrf2-antioxidant signalling. Cell Physiol Biochem. 2017;43:1140–1151. doi:https://doi.org/10.1159/000481755.
- Li P, Su L, Li X, et al. Remote limb ischemic postconditioning protects mouse brain against cerebral ischemia/reperfusion injury via upregulating expression of Nrf2, HO-1 and NQO-1 in mice. Int J Neurosci. 2015;126:1–8. doi:https://doi.org/10.3109/00207454.2015.1042973.
- Cheng Z, Li L, Mo X, et al. Non-invasive remote limb ischemic postconditioning protects rats against focal cerebral ischemia by upregulating STAT3 and reducing apoptosis. Int J Mol Med. 2014;34:957–966. doi:https://doi.org/10.3892/ijmm.2014.1873.
- Kim YH, Yoon DW, Kim JH, et al. Effect of remote ischemic post-conditioning on systemic inflammatory response and survival rate in lipopolysaccharide-induced systemic inflammation model. J Inflamm (Lond). 2014;11:16. doi:https://doi.org/10.1186/1476-9255-11-16.
- Li HX, Cui XL, Xue FS, et al. Inhibition of glycogen synthase kinase-3β is involved in cardioprotection by α7nAChR agonist and limb remote ischemic postconditionings. Biosci Rep. 2018;38. doi:https://doi.org/10.1042/BSR20181315.
- Mitchell TS, Moots RJ, Wright HL. Janus kinase inhibitors prevent migration of rheumatoid arthritis neutrophils towards interleukin-8, but do not inhibit priming of the respiratory burst or reactive oxygen species production. Clin Exp Immunol. 2017;189:250–258. doi:https://doi.org/10.1111/cei.12970.
- Levert H, Gressier B, Moutard I, et al. Azithromycin impact on neutrophil oxidative metabolism depends on exposure time. Inflammation. 1998;22:191–201. doi:https://doi.org/10.1023/a:1022340107017.
- el Benna J, Faust LP, Babior BM. The phosphorylation of the respiratory burst oxidase component p47phox during neutrophil activation. phosphorylation of sites recognized by protein kinase C and by proline-directed kinases. J Biol Chem. 1994;269:23431–23436.
- Cui YD, Inanami O, Yamamori T, et al. FMLP-induced formation of F-actin in HL60 cells is dependent on PI3-K but not on intracellular Ca2+, PKC, ERK or p38 MAPK. Inflamm Res. 2000;49:684–691. doi:https://doi.org/10.1007/s000110050647.
- Dang PM, Elbim C, Marie JC, et al. Anti-inflammatory effect of interleukin-10 on human neutrophil respiratory burst involves inhibition of GM-CSF-induced p47PHOX phosphorylation through a decrease in ERK1/2 activity. FASEB J. 2006;20:1504–1506. doi:https://doi.org/10.1096/fj.05-5395fje.
