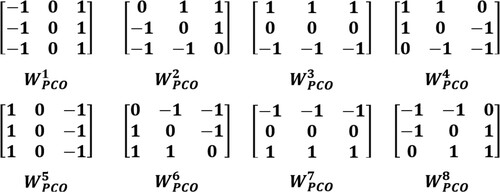?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Medical image fusion plays a pivotal role in facilitating clinical diagnosis. However, the quality of input medical images may be marred by noise, low contrast, and lack of sharpness, presenting numerous challenges for medical image synthesis algorithms. Additionally, several fusion rules may degrade the brightness and contrast of the fused image. To this end, this paper presents a novel image synthesis approach to tackle the aforementioned issues. First, the input images undergo pre-processing to enhance their quality. Subsequently, we introduce the three-layer image decomposition (TLID) technique, which decomposes an image into three distinct layers: the base layer (), the small-scale structure layer (
), and the large-scale structure layer (
). Next, we synthesize the base layers utilizing adaptive rules based on the Marine predators algorithm (MPA), ensuring that the output image is not degraded. Finally, we propose an efficient synthesis method for
and
layers, based on combining the local energy function with its variations. This fusion technique preserves the intricate details present in the original image. We evaluated our approach on 156 medical images using six evaluation metrics and compared it with seven state-of-the-art image synthesis techniques. Our results demonstrate that our method successfully generates high-quality output images and preserves detailed information throughout the image synthesis process.
1. Introduction
Medical imaging is an essential component of clinical applications today. With the advent of advanced image acquisition devices, a variety of multimodal medical images can now be obtained. Human organ structures are highly complex, and lesions cannot be adequately described using a single type of multimodal medical imaging. Computed Tomography (CT), magnetic resonance imaging (MRI), positron emission tomography (PET), and single photon emission computed tomography (SPECT) are among the commonly used types of multimodal medical imaging in image fusion. SPECT imaging is particularly useful for diagnosing vascular conditions and detecting tumors, as it offers insights into metabolic activity. In contrast, MRI is renowned for its ability to capture high-resolution images of soft tissues. To provide clinicians with comprehensive diagnostic information, it is crucial to synthesize the necessary information from multimodal medical images.
Traditional approaches to image fusion involve a sequence of three fundamental steps: image decomposition, synthesis of the decomposed components, and transforming the resulting fused image back to the original domain. Presently, various image decomposition methods exist. The first group of such methods comprises multi-scale decomposition (MSD)-based techniques, such as Discrete wavelet transform (DWT), Stationary wavelet transform (SWT), and Laplacian pyramid (LP). For example, Wang et al. [Citation1] utilized the DWT method to decompose input images before fusing them, while Dinh [Citation2] applied the SWT method to obtain high and low-frequency components of input images. Fu et al. [Citation3] employed the LP method to decompose input images. However, MSD-based methods have limited capacity to capture directional information, thus resulting in suboptimal image representation. The second group of methods is multi-scale geometric analysis (MGA)-based approaches, which overcome the limitations of MSD-based methods. These methods include curvelet transform, contourlet transform (CT), non-subsampled contourlet transform (NSCT), shearlet transform (ST), and non-subsampled shearlet transform (NSST). They provide complete information about both phase and direction. For example, Wang et al. [Citation4] employed NSCT to decompose input images, while Gao et al. [Citation5] utilized NSST to transform input images. Several other image synthesis studies based on MGA methods exist, including [Citation6], [Citation7], and [Citation8]. However, these methods have high computational complexity. The third group of image transformation methods comprises base-detail decomposition (BDD) based techniques, which exhibit benefits over MSD-based methods in terms of both computational efficiency and fusion performance. BDD-based methods enable the decomposition of an input image into base and detail layers by employing filters. For instance, Dinh [Citation9] proposed a novel image fusion approach based on two-scale image decomposition. Li et al. [Citation10] applied three-layer decomposition to fuse medical images. The fourth group of image transformation methods consists of Sparse representation (SR)-based methods, which have proven to be highly effective in various image synthesis studies. Wang et al. [Citation11] presented a new multi-focus image fusion approach based on multi-scale SR. Jie et al. [Citation12] integrated the SR with adaptive energy-choosing schemes to fuse Tri-modal medical images. Total-variational decomposition (TVD) has also been leveraged in several studies for image synthesis. Liu et al. [Citation13] proposed a novel image synthesis method based on the Robust spiking cortical model and TVD. Additionally, Liu et al. [Citation14] utilized spectral total variation and local structural patch measurement to construct an image composite model.
Traditional methods of image fusion have limitations that still persist. One limitation is that these methods are bound to use the same image decomposition method to acquire features and ensure their combinability in the subsequent phase. This results in the disregard of source image differences, which ultimately leads to poor expressiveness of the extracted features. Additionally, traditional methods acquire few and insufficiently diverse features from image decomposition, which still poses limitations on their synthesis performance. Deep learning has proven to be a powerful tool for addressing image processing problems, including image enhancement [Citation15], image denoising [Citation16], and image fusion. In particular, deep learning-based approaches have been instrumental in overcoming the limitations of traditional image fusion methods. Firstly, these approaches can conduct differentiated feature extraction by utilizing diverse network branches. Secondly, the feature fusion strategy can be effectively learned by designing appropriate loss functions. As a result, deep learning-based methods have significantly contributed to image fusion. For instance, Hou et al. [Citation17] proposed a novel method for synthesizing CT and MRI images by combining convolutional neural networks with a dual-channel spiking cortical model. Ding et al. [Citation18] presented an image synthesis method based on Dual-Branch CNNs in the NSST domain. Additionally, Ding et al. [Citation19] employed Siamese networks in conjunction with the multi-scale local extrema scheme. Wang et al. [Citation1] combined Convolutional neural networks (CNN) with Discrete wavelet transform (DWT) to fuse Multi-focus images. Kaur et al. [Citation20] proposed a medical image fusion based on deep belief networks (DBN). Other studies have also employed deep learning techniques to synthesize images, including [Citation21–25].
In recent times, meta-heuristic optimization-based image synthesis methods have been proven effective. Optimization algorithms offer adaptive rules for the synthesis process, leading to an enhanced quality of the output composited image. For example, Dinh [Citation26] proposed a new method that uses the Equilibrium optimizer algorithm (EOA) to synthesize medical images. Shilpa et al. [Citation27] improved the JAYA optimization algorithm and applied it to fuse medical images in the NSST domain. Particle swarm optimization (PSO) was applied by Shehanaz et al. [Citation28] to fuse high-frequency components in the DWT domain. Xu et al. [Citation29] modified the shark smell optimization (SSO) algorithm and combined it with the World Cup Optimization (WCO) algorithm to synthesize low-frequency components in the DWT domain. Dinh [Citation30] utilized the Chameleon swarm algorithm to synthesize the base components, ensuring the preservation of composite image quality. Further optimization techniques for producing medical images are discussed in the research papers [Citation9,Citation31–34].
Based on our observations, the low efficiency of image fusion can be attributed to three main factors. Firstly, low-quality input images are a common issue, characterized by low brightness and contrast, noise, and lack of sharpness, which negatively impacts the quality of the composite image. Secondly, the average rule, a widely adopted method in several studies [Citation35–37], has the advantage of simplicity and low computational complexity in synthesizing low-frequency components. However, its disadvantage is that it leads to a degradation in the brightness and contrast of the output composite image. Thirdly, the synthesis rules for high-frequency components have not been designed efficiently to capture full details from the input images, further impacting the quality of the composite image. In light of the aforementioned limitations, we propose the following approaches to overcome them. Firstly, we suggest enhancing the quality of input images using the Brighten low-light image (BLLI) method [Citation38]. Secondly, we propose using the MPA optimization algorithm to generate adaptive rules for low-frequency components, thereby ensuring optimal brightness and contrast in the output image. Thirdly, we introduce a fusion rule based on the local energy function and its variations to generate an efficient fusion rule for detail layers. This is because the local energy function and its variations have proven effective in many image synthesis studies. For instance, Amini et al. [Citation39] combined the local energy function with local variance fusion rules to synthesize MRI and PET images. Several variations of the local energy function have been proposed in recent years for medical image synthesis. Compass operators, such as Kirsch and Prewitt, have been combined with a local energy function to synthesize medical images [Citation9, Citation26]. The structure tensor has been applied in several studies on image fusion [Citation40–42]. Recently, Li et al. [Citation43] combined the structure tensor salient detection operator with a local energy function to construct a synthesis rule for detail components.
In this work, we present a novel method to tackle the aforementioned limitations. Our principal contributions are summarized as follows:
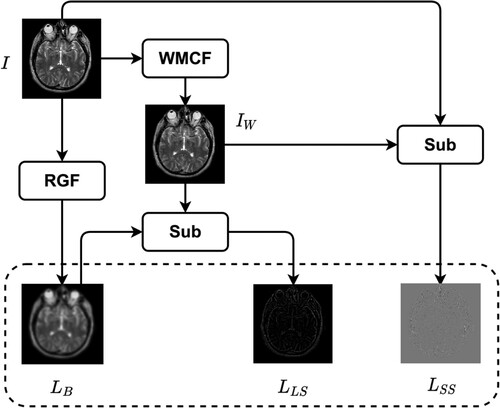
Firstly, The present study proposes a novel three-layer image composition (TLID) method for image decomposition into three layers, namely, the base layer (
), small-scale structure layer (
), and large-scale structure layer (
). This approach utilizes two filters, namely, the Rolling Guidance Filter (RGF) and the Weighted Median curvature filter (WMCF), to construct the TLID method.
Secondly, we propose an efficient fusion method to merge the
and
layers, which involves the combination of the local energy function with its variations.
Thirdly, in order to mitigate the loss of brightness and contrast during the image compositing process, we propose a novel method for fusing the base layers utilizing adaptive parameters.
The paper is structured as follows: Section 2 provides an overview of background methods including Rolling Guidance Filter (RGF), Weighted mean curvature filter (WMCF), Structure Tensor, Local Energy and its variations, and MPA algorithm. Section 3 introduces the TLID method, which combines variations of the local energy function (FR-CVLEF) to form a new fusion rule, as well as our proposed image fusion model. Section 4 contains experimental data and settings, results, and evaluations. Finally, Section 5 concludes the paper and presents future work.
2. Background
2.1. Rolling guidance filter (RGF)
The Rolling Guidance Filter (RGF) [Citation44] is a digital image filtering technique that has gained popularity in recent years due to its ability to effectively denoise images while preserving their sharpness and details. RGF is a non-local means filter that is designed to exploit the global structures and textures of an image, which allows it to effectively remove noise without blurring the image or losing important information. The filter comprises of two primary stages: removing small structures and recovering edges.
Step 1: The small structure is removed by the GF.
The symbols I and refer to the input and output images obtained following filtration via the Gaussian filter, respectively. The computation of
is performed in accordance with Equation (Equation1
(1)
(1) ).
(1)
(1) where
u and v are the position.
is the standard deviation of GF.
is the set of neighboring pixels whose center is u.
is calculated according to Equation (Equation2
(2)
(2) ).
The symbol denotes the GF output from the initial step.
represents the outcome achieved at the t-th iteration. Computation of
is executed in accordance with Equation (Equation3
(3)
(3) ).
(3)
(3) where
is calculated according to Equation (Equation4
(4)
(4) ).
is the standard deviation of the Guided Filter.
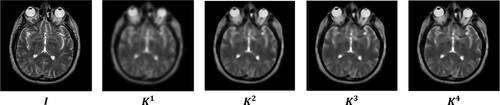
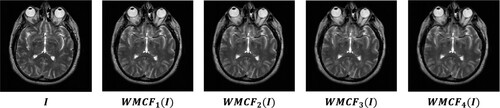
2.2. Weighted mean curvature filter (WMCF)
The WMCF is a technique used in image processing and computer vision. It was introduced by Gong et al. [Citation45], and its advantages are scale invariance, sampling invariance, and contrast invariance. Several applications of this filter can be mentioned as multi-spectral and panchromatic image fusion [Citation46], as well as medical image fusion [Citation47]. We denote an input image as R, and define the symbol (
) as the mask matrices of the WMFC filter, as illustrated in .
The WMCF can be calculated in two steps:
Step 1: Determine the distance as in Equation (Equation5
(5)
(5) ).
(5)
(5)
Step 2: This filter is defined as Equation (Equation6(6)
(6) ).
(6)
(6) where
illustrates the results of using the WMCF.
2.3. Structure tensor (ST)
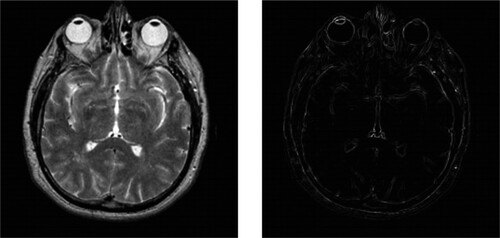
The computation of the ST is based on the gradient of the gray-scale image, and finds wide-ranging applications such as hyperspectral and panchromatic image fusion [Citation48], image denoising [Citation49], and medical image fusion [Citation43]. Let I be an input image. The ST can be calculated using Equation (Equation7(7)
(7) ).
(7)
(7) where,
w is a local window.
and
are the gradients in the i-direction and j-direction, respectively.
Furthermore, the operator for detecting salient features using the structure tensor, referred to as the Structure Tensor Salient Detection Operator (STSDO) [Citation50], is derived from the eigenvalues ( and
) as per Equation (Equation8
(8)
(8) ).
(8)
(8) Where
and
are determined according to Equations (Equation9
(9)
(9) ) and (Equation10
(10)
(10) ).
(9)
(9)
(10)
(10) illustrates the image obtained by applying the STSD operator.
2.4. Local energy function and its variations
2.4.1. Local energy function
The Local Energy Function (LEF) has found numerous applications in studies related to image fusion [Citation39, Citation51]. The computation of is performed using Equation (Equation11
(11)
(11) ).
(11)
(11)
is a unit window of size
.
I is the input image.
2.4.2. Local energy function using the Prewitt compass operator (LEF_PCO)
Dinh [Citation26] proposed the LEF-PCO method for developing the synthesis rule for detail components. We denote an input image as I, and refer to the k-th mask of the Prewitt compass operator as , with further clarification provided in . The computation of LEF-PCO is carried out using Equation (Equation12
(12)
(12) ).
(12)
(12)
2.4.3. Local energy function combined with the structure tensor saliency
Dinh [Citation52] introduced the Local Energy Function combined with the Structure Tensor Salient Detection Operator (LEF_STSDO) for generating fusion rules for detailed components.
The LEF_STSDO is defined as Equation (Equation13(13)
(13) ).
(13)
(13) where,
is defined as the STSDO in Equation (Equation8
(8)
(8) ).
⊙ represents entry-wise multiplication.
LEF represents the local energy function and is defined by Equation (Equation11
(11)
(11) ).
2.5. MPA algorithm
The MPA algorithm was initially proposed by Faramarzi et al. [Citation53]. This algorithm has exhibited superior optimization performance compared to other algorithms such as Genetic algorithm (GA), Gravitational Search Algorithm (GSA) [Citation54], and Salp Swarm Algorithm (SSA) [Citation55]. The MPA algorithm has found numerous applications in various domains such as structural damage detection [Citation56], image segmentation [Citation57], and medical image fusion [Citation52, Citation58]. The MPA algorithm can be outlined in three main steps as follows:
Step 1: During the first third of the loop, the prey exhibits faster movement than the predator. The calculation of stepsize () and prey (
) is performed using Equations (Equation14
(14)
(14) ) and (Equation15
(15)
(15) ).
(14)
(14)
(15)
(15) Where
in [0,1].
h = 0.5.
⊗ is entry-wise multiplication.
is selected randomly from a Brownian motion distribution.
holds the fitness solution.
Step 2: During the subsequent third of the loop, updates to and
are made using Equations (Equation16
(16)
(16) ), (Equation17
(17)
(17) ), (Equation18
(18)
(18) ), and (Equation19
(19)
(19) ).
For the first half of the population:
(16)
(16)
(17)
(17) For the second half of the population:
(18)
(18)
(19)
(19) where
.
is generated from the
distribution.
Step 3: Updates to and
are made during the final third of the loop using Equations (Equation20
(20)
(20) ) and (Equation21
(21)
(21) ).
(20)
(20)
(21)
(21) Fish Aggregating Devices (FDAs) effect:
is updated according to Equation (Equation22
(22)
(22) ).
(22)
(22) where,
represents the binary vector array.
k is a uniform random number in the range of [0,1].
and
represent random indexes of the prey matrix.
3. Our approach
This section presents three algorithms. The first algorithm is the TLID method. The second algorithm introduces a new fusion rule that combines variations of the local energy function. The third algorithm proposes our image fusion method.
3.1. Three-layer image decomposition method
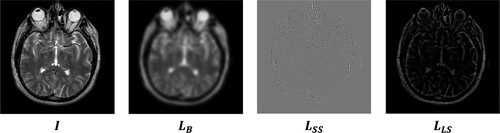
Image synthesis begins with image decomposition, which involves separating the source image into layers with complementary information. Typically, image decomposition algorithms create a base layer and one or more detail layers. In previous studies, a two-layer image decomposition method was commonly used, with the base layer obtained using the average [Citation59] or low-pass filters [Citation35]. However, these filters can cause loss of detailed information in the image, resulting in incomplete detail layers. To address these limitations, we propose a three-layer image decomposition method based on RGF and WMC filters. The algorithmic steps of this method are presented in Algorithm 1, and the process is illustrated in . An illustrative example of a three-layer image decomposition is illustrated in .
Algorithm 1. The three-layer image decomposition
3.2. Fusion rules based on combining variations of the local energy function
In this subsection, we introduce a novel fusion rule, named FR-CVLEF, which combines variations of the local energy function to fuse the small-scale and large-scale structure layers.
Algorithm 2. FR-CVLEF
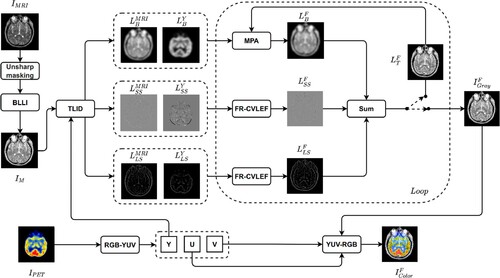
3.3. Our image fusion method
In this subsection, we present our approach, which involves three main steps. Firstly, we enhance the input image () using two methods: unsharp masking and the Brighten Low-Light Image (BLLI) method [Citation38]. Next, the image
is transformed into the channels Y, U, and V. We then decompose
and Y into base layers, small-scale structure layers (
), and large-scale structure layers (
) using the TLID method. After that, we fuse the base layers using an adaptive rule and the
and
layers using the FR-CVLEF to obtain the synthesized layers
,
and
, respectively. We then calculate the composite gray image (
) by summing the three layers,
,
and
. Finally, we convert
, U, and V into a color composite image (
). Our approach is illustrated in Algorithm 3 and .
Algorithm 3. The proposed approach
4. Experimental setup and evaluation
4.1. Experimental data
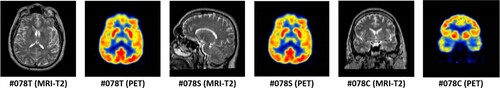
A total of 156 images, including 78 pairs of MRI and PET images, were utilized in this study. The images were sourced from ‘The Whole Brain Atlas’ (http://www.med.harvard.edu/AANLIB/) and are described in detail in . The three pairs of images in the K4 dataset are illustrated in .
Table 1. Dataset.
4.2. Experimental setup
We design some experiments as follows:
Experiment #1
We selected several optimization algorithms for comparison with the MPA algorithm. The algorithms considered are described in , and the experimental data used in this study is the K4 dataset.
Table 2. Five optimization algorithms.
Experiment #2
Several fusion rules were used for comparison with the proposed rule (FR_CVLEF), as outlined below:
Max selection rule (
).
Maximum local energy (
) [Citation63].
PA-PCNN (
) [Citation64].
Sum-modified laplacian (
) [Citation65].
Experiment #3
Our study involves a comparative analysis between our algorithm and seven contemporary image fusion algorithms, which are comprehensively described in .
Table 3. Seven image fusion algorithms.
We compare our algorithm with seven contemporary image fusion algorithms (as detailed in ).
For Experiments #2 and #3, we utilized data sets K1, K2, and K3. We evaluated the performance of the image fusion algorithms using the following five indicators:
Average light intensity (
).
Contrast index (
).
Sharpness (
).
Edge-based similarity measure (
) [Citation71].
Feature mutual information [Citation72] (
).
The necessary parameters used in our model are set as follows:
n = 50,
.
h = 0.5;
.
4.3. Results and evaluation
The results of the three experiments described in Section 4.2 are presented here. In the first experiment, we ran each optimization algorithm 30 times independently and evaluated the mean () and standard deviation (
) using two metrics. The results are presented in and . The MPA algorithm outperformed the others, producing the highest
value and the lowest
value. Therefore, it is the preferred choice for the proposed model. We also conducted a Wilcoxon rank-sum test [Citation73] to assess the significance of the results, and the P-values were found to be above 0.05, indicating statistical significance, as shown in .
Figure 11. The fitness function values obtained from various optimization algorithms in 30 independent on dataset K4 runs are visually presented in the form of a box plot.
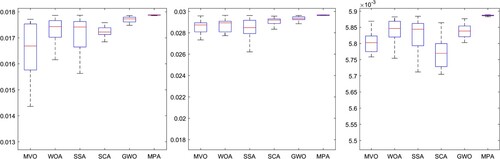
Table 4. and
from 30 different runs.
Table 5. P-values from Wilcoxon test.
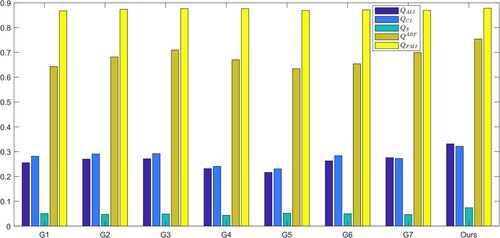
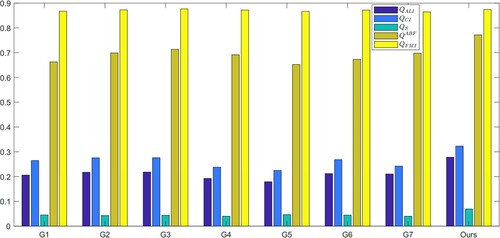
In the second experiment, we compared our fusion rules with four other fusion rules for SS and LS layers. We evaluated the five indicators, and the results are presented in . Our FR_CVLEF rule performed the best as it obtained the highest scores for two evaluation indicators, and
, compared to the other fusion rules.
Table 6. Two evaluation metrics ( and
) obtained from different fusion rules.
For the third experiment, adaptive parameters were obtained and are listed in . A value of approximately 1 for indicates that the fused image has the highest similarity to the MRI image. Moreover, the
value is considerably larger than the
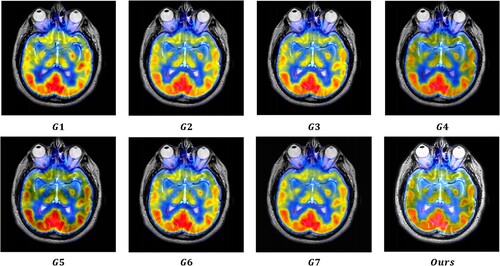
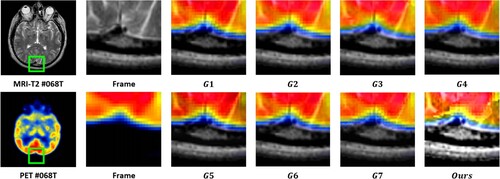
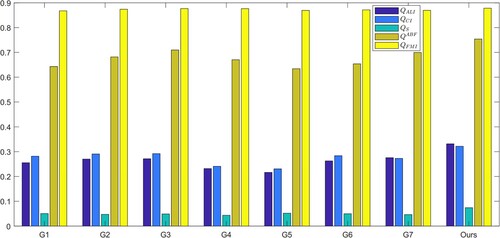
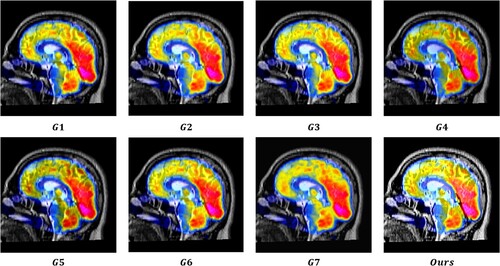
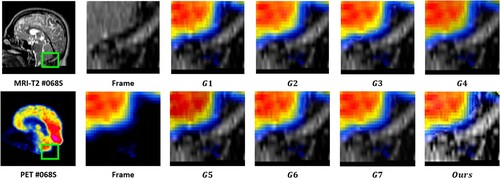
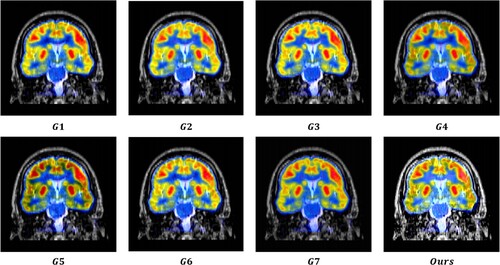
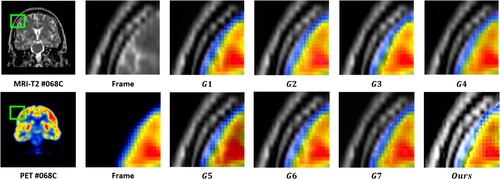
value, which suggests that the MRI images provide substantial information to the output image. The output images were generated using seven image fusion algorithms, including our own algorithm, which is depicted in Figures , , and . Visual inspection indicates that the output image produced by our algorithm exhibits high quality in terms of light intensity, contrast, and sharpness. A small area was cropped from the image to observe its details, as shown in Figures , , and . The details in these small areas are well preserved. In terms of quantitative analysis, presents the five evaluation indicators obtained from various image fusion algorithms. The proposed method's output image has significantly higher quality than the other image fusion methods due to the adaptive rule used for the base layers. The FR_CVLRF method guarantees the output images retain the input image details.
Table 7. Adaptive parameters obtained in three data sets K1, K2, and K3.
Table 8. The evaluation indicators obtained from image fusion algorithms.
displays the average running times of the image fusion algorithms. The algorithm developed by us takes an average of 4.58 seconds to complete the fusion process. However, this time increases as the number of iterations increases, making it a disadvantage of our algorithm.
Table 9. Average running time (ART) of image fusion algorithms on the data set (K1&K2&K3).
5. Conclusion
In this paper, a new approach for fusing medical images is introduced, which combines a local energy function and variations of it with the MPA algorithm. The proposed method starts by introducing a three-layer image decomposition technique, which separates the input images into three distinct layers: the base layer (), the small-scale structure layer (
), and the large-scale structure layer (
). Next, a fusion rule built on combining the local energy function with its variations is introduced to fuse the
and
layers. Thirdly, the base layers are synthesized based on the MPA algorithm. The method was tested using 156 MRI-T2 and PET images, along with five evaluation metrics and seven state-of-the-art image fusion algorithms. The proposed adaptive rules for the base layers resulted in an output composite image that had a high level of quality in terms of brightness intensity and contrast. For example, from , the
and
indexes obtained from the proposed model are the highest, at 0.3131 and 0.3356 on data set K1. Furthermore, applying the FR_CVLEF method to the small-scale and large-scale structure layers has also shown significant efficiency in the detailed information of the output image. For instance, from , the
,
, and
indexes obtained from the proposed model are the highest, at 0.0829, 0.7440, and 0.8737 on data set K1, respectively.
In the future, our goal is to enhance the performance of the proposed model in several aspects. First, apply new image enhancement methods [Citation74–76] to replace the BLLI method. This allows for significantly improved performance of image synthesis. Second, select some recent optimization algorithms, such as the Chameleon swarm algorithm (CSA) [Citation77, Citation78] and White Shark Optimizer (WSO) [Citation79], to replace the MPA algorithm. This can improve the runtime of the proposed model.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Notes on contributors
Phu-Hung Dinh
Phu-Hung Dinh was born in Vietnam. Currently, he is a lecturer at the Faculty of Computer Science and Engineering, Thuyloi University, Hanoi, Vietnam. His research interests include signal processing, image processing, meta-heuristic optimization, and computer vision. Furthermore, he is also a reviewer for many prestigious journals, such as Expert systems with applications, Artificial intelligence review, Computers in biology and medicine, Biomedical signal processing & control, ISA transactions, BMC Bioinformatics, Applied Intelligence, Soft Computing, Imaging science journal, and Journal of Supercomputing.
References
- Wang Z, Li X, Duan H, et al. Multifocus image fusion using convolutional neural networks in the discrete wavelet transform domain. Multimed Tools Appl. 2019;78(24):34483–34512. doi:10.1007/s11042-019-08070-6.
- Dinh PH. Combining gabor energy with equilibrium optimizer algorithm for multi-modality medical image fusion. Biomed Signal Process Control. 2021;68:102696. doi:10.1016/j.bspc.2021.102696.
- Fu J, Li W, Du J, et al. Multimodal medical image fusion via laplacian pyramid and convolutional neural network reconstruction with local gradient energy strategy. Comput Biol Med. 2020;126:104048. doi:10.1016/j.compbiomed.2020.104048.
- Wang Z, Li X, Duan H, et al. Medical image fusion based on convolutional neural networks and non-subsampled contourlet transform. Expert Syst Appl. 2021;171:114574. doi:10.1016/j.eswa.2021.114574.
- Gao Y, Ma S, Liu J, et al. Fusion of medical images based on salient features extraction by PSO optimized fuzzy logic in NSST domain. Biomed Signal Process Control. 2021;69:102852. doi:10.1016/j.bspc.2021.102852.
- Nair RR, Singh T. MAMIF: multimodal adaptive medical image fusion based on b-spline registration and non-subsampled shearlet transform. Multimed Tools Appl. 2021;80(12):19079–19105. doi:10.1007/s11042-020-10439-x.
- Wang S, Shen Y. Multi-modal image fusion based on saliency guided in NSCT domain. IET Image Process. 2020;14:3188–3201. doi:10.1049/iet-ipr.2019.1319.
- Nair RR, Singh T. An optimal registration on shearlet domain with novel weighted energy fusion for multi-modal medical images. Optik. 2021;225:165742. doi:10.1016/j.ijleo.2020.165742.
- Dinh PH. A novel approach based on grasshopper optimization algorithm for medical image fusion. Expert Syst Appl. 2021;171:114576. doi:10.1016/j.eswa.2021.114576.
- Li X, Zhou F, Tan H. Joint image fusion and denoising via three-layer decomposition and sparse representation. Knowl Based Syst. 2021;224:107087. doi:10.1016/j.knosys.2021.107087.
- Wang Y, Li X, Zhu R, et al. A multi-focus image fusion framework based on multi-scale sparse representation in gradient domain. Signal Processing. 2021;189:108254. doi:10.1016/j.sigpro.2021.108254.
- Jie Y, Zhou F, Tan H, et al. Tri-modal medical image fusion based on adaptive energy choosing scheme and sparse representation. Measurement. 2022;204:112038. doi:10.1016/j.measurement.2022.112038.
- Liu Y, Zhou D, Nie R, et al. Robust spiking cortical model and total-variational decomposition for multimodal medical image fusion. Biomed Signal Process Control. 2020;61:101996. doi:10.1016/j.bspc.2020.101996.
- Liu Y, Hou R, Zhou D, et al. Multimodal medical image fusion based on the spectral total variation and local structural patch measurement. Int J Imaging Syst Technol. 2020;31(1):391–411. doi:10.1002/ima.22460.
- Wang J, Li X, Wang Z, et al. Exposure correction using deep learning. J Electron Imaging. 2019;28(3):1. doi:10.1117/1.jei.28.3.033003.
- Gai S, Bao Z. New image denoising algorithm via improved deep convolutional neural network with perceptive loss. Expert Syst Appl. 2019;138:112815. doi:10.1016/j.eswa.2019.07.032.
- Hou R, Zhou D, Nie R, et al. Brain CT and MRI medical image fusion using convolutional neural networks and a dual-channel spiking cortical model. Med Biol Eng Comput. 2018;57(4):887–900. doi:10.1007/s11517-018-1935-8.
- Ding Z, Zhou D, Nie R, et al. Brain medical image fusion based on dual-branch CNNs in NSST domain. Biomed Res Int. 2020;2020:1–15. doi:10.1155/2020/6265708.
- Ding Z, Zhou D, Li H, et al. Siamese networks and multi-scale local extrema scheme for multimodal brain medical image fusion. Biomed Signal Process Control. 2021;68:102697. doi:10.1016/j.bspc.2021.102697.
- Kaur M, Singh D. Fusion of medical images using deep belief networks. Cluster Comput. 2019;23(2):1439–1453. doi:10.1007/s10586-019-02999-x.
- Yousif AS, Omar Z, Sheikh UU. An improved approach for medical image fusion using sparse representation and siamese convolutional neural network. Biomed Signal Process Control. 2022;72:103357. doi:10.1016/j.bspc.2021.103357.
- Guo K, Hu X, Li X. MMFGAN: A novel multimodal brain medical image fusion based on the improvement of generative adversarial network. Multimed Tools Appl. 2021;81:5889–5927. doi:10.1007/s11042-021-11822-y.
- Li W, Peng X, Fu J, et al. A multiscale double-branch residual attention network for anatomical–functional medical image fusion. Comput Biol Med. 2021;141:105005. doi:10.1016/j.compbiomed.2021.105005.
- Wang Z, Li X, Yu S, et al. VSP-fuse: multifocus image fusion model using the knowledge transferred from visual salience priors. IEEE Trans Circuits Syst Video Technol. 2022:1–1. doi:10.1109/tcsvt.2022.3229691.
- Wang Z, Li X, Duan H, et al. A self-supervised residual feature learning model for multifocus image fusion. IEEE Trans Image Process. 2022;31:4527–4542. doi:10.1109/tip.2022.3184250.
- Dinh PH. Multi-modal medical image fusion based on equilibrium optimizer algorithm and local energy functions. Appl Intell. 2021;51:8416–8431. doi:10.1007/s10489-021-02282-w.
- Shilpa S, Rajan MR, Asha C, et al. Enhanced JAYA optimization based medical image fusion in adaptive non subsampled shearlet transform domain. Eng Sci Technol Int J. 2022;35:101245. doi:10.1016/j.jestch.2022.101245.
- Shehanaz S, Daniel E, Guntur SR, et al. Optimum weighted multimodal medical image fusion using particle swarm optimization. Optik. 2021;231:166413. doi:10.1016/j.ijleo.2021.166413.
- Xu L, Si Y, Jiang S, et al. Medical image fusion using a modified shark smell optimization algorithm and hybrid wavelet-homomorphic filter. Biomed Signal Process Control. 2020;59:101885. doi:10.1016/j.bspc.2020.101885.
- Dinh PH. Medical image fusion based on enhanced three-layer image decomposition and chameleon swarm algorithm. Biomed Signal Process Control. 2023;84:104740. doi:10.1016/j.bspc.2023.104740.
- Jose J, Gautam N, Tiwari M, et al. An image quality enhancement scheme employing adolescent identity search algorithm in the NSST domain for multimodal medical image fusion. Biomed Signal Process Control. 2021;66:102480. doi:10.1016/j.bspc.2021.102480.
- Dinh PH. An improved medical image synthesis approach based on marine predators algorithm and maximum gabor energy. Neural Comput Appl. 2021;34:4367–4385. doi:10.1007/s00521-021-06577-4.
- Nguyen TT, Wang HJ, Dao TK, et al. A scheme of color image multithreshold segmentation based on improved moth-flame algorithm. IEEE Access. 2020;8:174142–174159. doi:10.1109/access.2020.3025833.
- Nguyen TT, Ngo TG, Dao TK, et al. Microgrid operations planning based on improving the flying sparrow search algorithm. Symmetry. 2022;14(1):168. doi:10.3390/sym14010168.
- Liu Y, Chen X, Ward RK, et al. Image fusion with convolutional sparse representation. IEEE Signal Process Lett. 2016;23(12):1882–1886. doi:10.1109/lsp.2016.2618776.
- Liu Y, Chen X, Ward RK, et al. Medical image fusion via convolutional sparsity based morphological component analysis. IEEE Signal Process Lett. 2019;26:485–489. doi:10.1109/lsp.2019.2895749.
- Maqsood S, Javed U. Multi-modal medical image fusion based on two-scale image decomposition and sparse representation. Biomed Signal Process Control. 2020;57:101810. doi:10.1016/j.bspc.2019.101810.
- Dong X, Wang G, Pang Y, et al. Fast efficient algorithm for enhancement of low lighting video. In: 2011 IEEE International Conference on Multimedia and Expo. IEEE; 2011. doi:10.1109/icme.2011.6012107.
- Amini N, Fatemizadeh E, Behnam H. MRI-PET image fusion based on NSCT transform using local energy and local variance fusion rules. J Med Eng Technol. 2014;38(4):211–219. doi:10.3109/03091902.2014.904014.
- Yin H. Tensor sparse representation for 3-d medical image fusion using weighted average rule. IEEE Trans Biomed Eng. 2018;65(11):2622–2633. doi:10.1109/tbme.2018.2811243.
- Zhou J, Xing X, Yan M, et al. A fusion algorithm based on composite decomposition for PET and MRI medical images. Biomed Signal Process Control. 2022;76:103717. doi:10.1016/j.bspc.2022.103717.
- Du J, Li W, Tan H. Three-layer medical image fusion with tensor-based features. Inf Sci (Ny). 2020;525:93–108. doi:10.1016/j.ins.2020.03.051.
- Li X, Zhou F, Tan H, et al. Multimodal medical image fusion based on joint bilateral filter and local gradient energy. Inf Sci (Ny). 2021;569:302–325. doi:10.1016/j.ins.2021.04.052.
- Zhang Q, Shen X, Xu L, et al. Rolling guidance filter. In: Computer Vision–ECCV 2014. Springer International Publishing; 2014. p. 815–830. doi:10.1007/978-3-319-10578-9_53.
- Gong Y, Goksel O. Weighted mean curvature. Signal Processing. 2019;164:329–339. doi:10.1016/j.sigpro.2019.06.020.
- Pan Y, Liu D, Wang L, et al. A multispectral and panchromatic images fusion method based on weighted mean curvature filter decomposition. Appl Sci. 2022;12(17):8767. doi:10.3390/app12178767.
- Tan W, Thitøn W, Xiang P, et al. Multi-modal brain image fusion based on multi-level edge-preserving filtering. Biomed Signal Process Control. 2021;64:102280. doi:10.1016/j.bspc.2020.102280.
- Dong W, Xiao S, Liang J, et al. Fusion of hyperspectral and panchromatic images using structure tensor and matting model. Neurocomputing. 2020;399:237–246. doi:10.1016/j.neucom.2020.02.050.
- Liu K, Xu W, Wu H, et al. Weighted hybrid order total variation model using structure tensor for image denoising. Multimed Tools Appl. 2022;82:927–943. doi:10.1007/s11042-022-12393-2.
- Zhou Z, Li S, Wang B. Multi-scale weighted gradient-based fusion for multi-focus images. Inf Fusion. 2014;20:60–72. doi:10.1016/j.inffus.2013.11.005.
- Polinati S, Dhuli R. Multimodal medical image fusion using empirical wavelet decomposition and local energy maxima. Optik. 2020;205:163947. doi:10.1016/j.ijleo.2019.163947.
- Dinh PH. A novel approach using structure tensor for medical image fusion. Multidimens Syst Signal Process. 2022;33(3):1001–1021. doi:10.1007/s11045-022-00829-9.
- Faramarzi A, Heidarinejad M, Mirjalili S, et al. Marine predators algorithm: A nature-inspired metaheuristic. Expert Syst Appl. 2020;152:113377. doi:10.1016/j.eswa.2020.113377.
- Rashedi E, Nezamabadi-pour H, Saryazdi S. GSA: A gravitational search algorithm. Inf Sci (Ny). 2009;179(13):2232–2248. doi:10.1016/j.ins.2009.03.004.
- Mirjalili S, Gandomi AH, Mirjalili SZ, et al. Salp swarm algorithm: A bio-inspired optimizer for engineering design problems. Adv Eng Softw. 2017;114:163–191. doi:10.1016/j.advengsoft.2017.07.002.
- Ho LV, Nguyen DH, Mousavi M, et al. A hybrid computational intelligence approach for structural damage detection using marine predator algorithm and feedforward neural networks. Comput Struct. 2021;252:106568. doi:10.1016/j.compstruc.2021.106568.
- Houssein EH, Hussain K, Abualigah L, et al. An improved opposition-based marine predators algorithm for global optimization and multilevel thresholding image segmentation. Knowl Based Syst. 2021;229:107348. doi:10.1016/j.knosys.2021.107348.
- Dinh PH. A novel approach based on three-scale image decomposition and marine predators algorithm for multi-modal medical image fusion. Biomed Signal Process Control. 2021;67:102536. doi:10.1016/j.bspc.2021.102536.
- Li S, Kang X, Hu J. Image fusion with guided filtering. IEEE Trans Image Process. 2013;22(7):2864–2875. doi:10.1109/tip.2013.2244222.
- Mirjalili S, Mirjalili SM, Hatamlou A. Multi-verse optimizer: a nature-inspired algorithm for global optimization. Neural Comput Appl. 2015;27(2):495–513. doi:10.1007/s00521-015-1870-7.
- Mirjalili S, Lewis A. The whale optimization algorithm. Adv Eng Softw. 2016;95:51–67. doi:10.1016/j.advengsoft.2016.01.008.
- Mirjalili S. SCA: a sine cosine algorithm for solving optimization problems. Knowl Based Syst. 2016;96:120–133. doi:10.1016/j.knosys.2015.12.022.
- Lu H, Zhang L, Serikawa S. Maximum local energy: an effective approach for multisensor image fusion in beyond wavelet transform domain. Comput Math Appl. 2012;64(5):996–1003. doi:10.1016/j.camwa.2012.03.017.
- Yin M, Liu X, Liu Y, et al. Medical image fusion with parameter-adaptive pulse coupled neural network in nonsubsampled shearlet transform domain. IEEE Trans Instrum Meas. 2019;68:49–64. doi:10.1109/tim.2018.2838778.
- Li X, Zhang X, Ding M. A sum-modified-laplacian and sparse representation based multimodal medical image fusion in laplacian pyramid domain. Med Biol Eng Comput. 2019;57(10):2265–2275. doi:10.1007/s11517-019-02023-9.
- Zhu Z, Zheng M, Qi G, et al. A phase congruency and local laplacian energy based multi-modality medical image fusion method in NSCT domain. IEEE Access. 2019;7:20811–20824. doi:10.1109/access.2019.2898111.
- Sufyan A, Imran M, Shah SA, et al. A novel multimodality anatomical image fusion method based on contrast and structure extraction. Int J Imaging Syst Technol. 2021;32(1):324–342. doi:10.1002/ima.22649.
- Li B, Peng H, Luo X, et al. Medical image fusion method based on coupled neural p systems in nonsubsampled shearlet transform domain. Int J Neural Syst. 2020;31(01):2050050. doi:10.1142/s0129065720500501.
- Li B, Peng H, Wang J. A novel fusion method based on dynamic threshold neural p systems and nonsubsampled contourlet transform for multi-modality medical images. Signal Processing. 2021;178:107793. doi:10.1016/j.sigpro.2020.107793.
- Zhu R, Li X, Huang S, et al. Multimodal medical image fusion using adaptive co-occurrence filter-based decomposition optimization model. Bioinformatics. 2021;38(3):818–826. doi:10.1093/bioinformatics/btab721.
- Xydeas C, Petrovic V. Objective image fusion performance measure. Electron Lett. 2000;36:308. doi:10.1049/el:20000267.
- Haghighat MBA, Aghagolzadeh A, Seyedarabi H. A non-reference image fusion metric based on mutual information of image features. Comput Electr Eng. 2011;37:744–756. doi:10.1016/j.compeleceng.2011.07.012.
- Wilcoxon F. Individual comparisons by ranking methods. Biometrics Bulletin. 1945;1(6):80. doi:10.2307/3001968.
- Su Y, Li Z, Yu H, et al. Multi-band weighted lp norm minimization for image denoising. Inf Sci (Ny). 2020;537:162–183. doi:10.1016/j.ins.2020.05.049.
- Dinh PH, Giang NL. A new medical image enhancement algorithm using adaptive parameters. Int J Imaging Syst Technol. 2022;32:2198–2218. doi:10.1002/ima.22778.
- Dinh PH. A novel approach based on marine predators algorithm for medical image enhancement. Sens Imaging. 2023;24(1):6. doi:10.1007/s11220-023-00411-y.
- Braik MS. Chameleon swarm algorithm: a bio-inspired optimizer for solving engineering design problems. Expert Syst Appl. 2021;174:114685. doi:10.1016/j.eswa.2021.114685.
- Dinh PH. Combining spectral total variation with dynamic threshold neural p systems for medical image fusion. Biomed Signal Process Control. 2023;80:104343. doi:10.1016/j.bspc.2022.104343.
- Braik M, Hammouri A, Atwan J, et al. White shark optimizer: A novel bio-inspired meta-heuristic algorithm for global optimization problems. Knowl Based Syst. 2022;243:108457. doi:10.1016/j.knosys.2022.108457.