Abstract
Objective
Next Generation Sequencing (NGS) is increasingly used for the diagnosis of rare genetic disorders. The aim of this study is to review the different approaches for economic evaluations of Next Generation Sequencing (NGS) in pediatric care used to date, to identify all costs, effects, and time horizons taken into account.
Methods
A systematic literature review was conducted to identify published economic evaluations of NGS applications in pediatric diagnostics, i.e. exome sequencing (ES) and/or genome sequencing (GS). Information regarding methodological approach, costs, effects, and time horizon was abstracted from these publications.
Results
Twenty-eight economic evaluations of ES/GS within pediatrics were identified. Costs included were mainly restricted to direct in-hospital healthcare costs and varied widely in inclusion of sort of costs and time-horizon. Nineteen studies included diagnostic yield and eight studies included cost-effectiveness as outcome measures. Studies varied greatly in terms of included sort of costs data, effects, and time horizon.
Conclusion
Large differences in inclusion of cost and effect parameters were identified between studies. Validity of outcomes can therefore be questioned, which hinders valid comparison and widespread generalization of conclusions. In addition to current health economic guidance, specific guidance for evaluations in pediatric care is therefore necessary to improve the validity of outcomes and furthermore facilitate comparable decision-making for implementing novel NGS-based diagnostic modalities in pediatric genetics and beyond.
Introduction
Worldwide, approximately 350 million people are affected by rare disorders, of which 50% had an onset during childhood and 80% have a genetic originCitation1,Citation2. The diagnosis of rare genetic disorders in children is challenging and time consuming due to, among other things, the rarity of the individual disorder, the variability of the clinical manifestations, the genetic heterogeneity, and the deficiencies in laboratory testing. Recent developments in Next Generation Sequencing (NGS) have made it possible to investigate all protein-coding regions or even entire genomes in one single time, i.e. by implementing exome sequencing (ES) or genome sequencing (GS), respectivelyCitation3. These developments have resulted in an increase in genetic diagnoses and a shortened time-to-diagnosis for patients with expected genetic disordersCitation4,Citation5.
These rapid technological developments are often of clinical relevance, and subsequently studies on economic impact are increasingly being performed to assess the (added) value of new diagnostic modalities. To ensure valid decision-making and high-quality studies, it is essential that these studies adhere to respective health economic guidelines. Unfortunately, a recent study has indicated that there is currently a lack of adherence to these “basic health economic” guidelines in economic evaluations of pediatric geneticsCitation6.
Unfortunately, adherence to these “basic health economic” guidelines does not 100% ensure the validity and quality of evaluations. Recent studies in other areas have indicated the need for additional, so-called disease-specific guidance, in order to ensure right choices are made on a disease level with regard to inclusion of cost data, outcome parameters, and length of the analysisCitation6. In addition to adherence to existing guidance it is therefore also essential to adhere and or to include (newly) developed guidance published in scientific journals, guidance which increases disease specific uniformity of methods and outcome measures. This would not only facilitate the ability to share and combine health economic data, but would possibly also lead to a decrease in research waste. An excellent example of such disease-specific guidance is the publication of Buchanan et al.Citation7, in which they outline characteristics which should be included in economic evaluations.
Moreover, next to a complete and comparable collection of costs and adherence to existing guidelines, there is ongoing discussion about the inclusion of effect measures for genetic technologies. In the field of health economics, combining costs and effects into one outcome measure, such as cost per life year gained or cost per quality adjusted life year gained, is routinely used and subsequently decision-makers need to decide on reimbursementCitation8. However, using a uniform effect measure, i.e. an outcome measurement which makes it easier to compare or combine studies, is very challenging within the field of pediatric genetics, as, most often, obtaining a genetic diagnosis does not immediately lead to treatment options.
Although recent reviews have indicated that challenges with comparability are prevailing in economic evaluations of pediatric genetics, they did not outline whether differences in included costs and effects are prevalent and which costs and effects were included by the individual studiesCitation6,Citation9. Also assessment on inclusion of disease-specific recommendations is often lacking.
An overview of all parameters used so far within previous economic evaluations is currently lacking. We therefore aim to review all economic evaluations of ES and/or GS performed for pediatric onset genetic disorders, identifying all costs, effects, and time horizon (i.e. time over which data was collected) included, and providing the differences in approaches taken. Approaches are compared to disease-specific guidance, as recently recommendedCitation7,Citation10.
Methods
Study design and search strategy
A systematic literature review was conducted (14 February 2021) to identify published economic evaluations, in which costs and effects are compared (model-based, prospective, and retrospective) of the clinical applications of ES and/or GS as a diagnostic tool for rare genetic disorders in a pediatric setting. Hereto, the following search strategy was applied: (sequence analysis OR high throughput nucleotide sequencing OR next generation sequencing OR whole genome sequencing OR whole exome sequencing) AND (costs and cost analysis OR cost-effectiveness) AND (children OR infant OR pediatric OR paediatric). The databases used for the search were PubMed/MEDLINE and EMBASE.
Two independent reviewers validated the published articles based on the following inclusion criteria: (i) the full-version of the article was available; (ii) the article was published in English; (iii) ES and/or GS are part of the economic evaluation described; and (iv) the study population consists of children aged 18 years or younger. The independent reviewers participated in both data extraction and article screening. Both reviewers independently selected studies suitable for data analysis based on the inclusion criteria. Discrepancies were resolved by discussing the article in question. There were no restrictions considering year of publication, since the role of NGS in genetic diagnostics (ES and/or GS) has only become more evident since 2009. Of all included studies, reference lists were reviewed to identify additional studies.
Data extraction
Characteristics including study population, health condition, sample size, comparison of genetic tests, time horizon, type of included costs (for example costs related to diagnostic testing or non-medical costs) and effects (i.e. outcome measures) were extracted and summarized to create an overview of all economic evaluations included in this study. Time horizon was defined as the duration over which costs and effects were included. For each included study, the final conclusion was extracted.
Data analysis
The included costs were compared to the cost components as stated by Drummond et al.Citation10 and Buchanan et al.Citation7 to determine which aspects are currently missing in the evaluations. Drummond et al.Citation10 outlined that the following costs should be included: (i) costs within the healthcare sector, consisting of all medical costs directly resulting from the intervention and costs incurred during life years gained; (ii) costs for the patient and family, for example travel expenses, own contributions, time spending costs (e.g. time spent on informal care, time needed to undergo treatment or lost working hours due to an intervention) or costs of informal care; and (iii) costs in other sectors, which can be costs incurred in sectors outside the healthcare system, for municipal services, education, or voluntary work.
Buchanan et al.Citation7 defined eight cost components, which were specifically attributable to the evaluation of genomic technologies: costs related to (i) patient recruitment (e.g. publicity and education of patients); (ii) blood or tissue sample collection; (iii) sample testing; (iv) data analysis; (v) communication of test results; (vi) actions taken based on tests results; (vii) training and infrastructure (e.g. costs related to staff training); and (viii) indirect costs. In order to judge which effects should be measured during an economic evaluation, an overview of all included effects was created.
Results
Studies identified
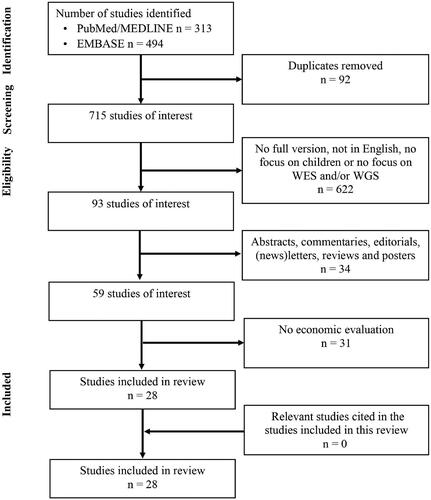
Based on our database searches, we identified 313 studies in Pubmed/MEDLINE and 494 studies in EMBASE, resulting in 807 unique studies. Of these, 28 studies (3.5%) fulfilled our inclusion criteria. Based on the PRISMA reporting guidelinesCitation11, an overview of the complete selection procedure is shown in . Manual inspection of the reference lists from these 28 studies did not yield any new studies.
In , the characteristics of the included studies are summarized. The rare disorders of the children for which the child received ES/GS (i.e. intellectual disability, epilepsy, autism spectrum disorder) and the cohort sizes varied (IQR = 40–300, median = 101). Nine studies (32.1%) included a scenario analysisCitation13,Citation16,Citation18,Citation23–26,Citation28,Citation37 and six (21.4%) investigated the implementation of GS instead of, or in addition to, ESCitation12,Citation21,Citation24,Citation26,Citation36,Citation39. Time horizons were also different for the included studies, and there was no consensus regarding the moment a study started nor the moment a study ended (). In more detail, five studies (17.9%) had a time horizon of a fixed duration (range = 7–24 months)Citation12,Citation23,Citation24,Citation29,Citation38 and six studies (21.4%) included the complete diagnostic trajectoryCitation16,Citation25,Citation31,Citation35–37. There were eight studies (30.8%) which started at a certain moment in time (i.e. first visit in hospital, onset of symptoms or moment of inclusion) until a diagnosis was foundCitation13,Citation15,Citation19,Citation20,Citation22,Citation28,Citation30,Citation32. Five studies (17.9%) looked at what had happened after ES was performedCitation21,Citation26,Citation27,Citation34,Citation39 and the remaining four studies (15.4%) included a time period from the onset of symptoms or first visit to the hospital until ES was initiatedCitation14,Citation17,Citation18,Citation33. The final conclusions of the included studies are shown in .
Table 1. Overview of characteristics of studies included in this review.
Included costs and effects
shows a summary of the different cost categories according to both Drummond et al.Citation10 and Buchanan et al.Citation7 Regarding inclusion of internal costs (i.e. diagnostic and non-medical costs within the hospital of interest), external costs (i.e. costs outside or in another hospital), and additional costs, results are diverse: 14 studies (50.0%) included only internal costsCitation12,Citation14,Citation15,Citation18,Citation24,Citation25,Citation29,Citation31–33,Citation35,Citation37–39, whereas 11 studies (42.3%) also took a part of the external costs into accountCitation13,Citation16,Citation17,Citation19–22,Citation27,Citation28,Citation30,Citation34; two studies (7.7%) also investigated nonmedical costs like traveling costsCitation23,Citation36 and time spending costs for (parents of) the patient, such as time lost due to medical visitsCitation26,Citation36. Furthermore, 11 studies (42.3%) solely focused on costs of diagnostic testingCitation12,Citation14,Citation16,Citation17,Citation19,Citation20,Citation22,Citation24,Citation25,Citation32,Citation35. None of the studies took costs incurred during life years gained or costs related to other sectors (non-medical costs) into account.
Table 2. Included costs categories according to Drummond et al.Citation10 and Buchanan et al.Citation7.
Compared to the cost components of Buchanan et al.Citation7, all of the studies included costs related to sample collection, sample testing, and data analysis. Fourteen out of 28 studies (50.0%) took post-test counseling into accountCitation13,Citation15,Citation18,Citation21,Citation23,Citation26,Citation27,Citation29,Citation31,Citation33,Citation36–39 and nine out of 28 studies (32.1%) took action taken based on test result into accountCitation13,Citation15,Citation18,Citation21,Citation23,Citation24,Citation27,Citation29,Citation39. Three studies (11.5%) included non-medical costs, such as travel expenses and time spending costs of patients and familyCitation23,Citation26,Citation36. Costs related to patient recruitment and training and infrastructure were not investigated.
shows a summary of the investigated effects. Nineteen studies (67.9%) included diagnostic yieldCitation12–14,Citation17–20,Citation22,Citation23,Citation25,Citation27,Citation29,Citation31,Citation32,Citation34,Citation35,Citation37–39. Two studies (7.7%) took the number of ongoing pregnancies and utilization of reproductive genetic services into accountCitation27,Citation34. Considering cost-related outcomes, included outcome effects varied: five studies (19.2%) focused on cost-effectiveness onlyCitation12,Citation14,Citation19,Citation22,Citation23, but the more recent publications included incremental costs per additional positive finding in (hypothetical) testing scenariosCitation24,Citation25, incremental costs per additional diagnosisCitation26–28,Citation31, or an incremental cost-effectiveness ratio (ICER)Citation34.
Table 3. Effect measures of the studies included in this review.
Discussion
Based on our inclusion criteria, to date 28 economic evaluations regarding NGS have been performed within the pediatric population. These economic evaluations were published between 2014 and 2020. Our review outlined the presence of an extensive variability in choice and usage of costs, effects, and time horizon in these economic evaluations. According to the cost categories defined by Drummond et al.Citation10, costs included by the studies mainly focused on diagnostic costs. Only three out of 28 took other healthcare resources (i.e. travel expensesCitation23,Citation36 and time spending costsCitation26,Citation36) into account, while personal costs (i.e. costs of informal care, own contributions/co-payments, and non-medical costs) were not included at all. The cost categories as defined by Drummond et al.Citation10 and Buchanan et al.Citation7 focus on the costs related to (genetic) diagnostics and beyond diagnostics. However, no uniformity can be found between the cost taken into account by the different studies in this review. In order to increase uniformity, future studies should, at a minimum, follow their guidelinesCitation7,Citation10. Since it will be challenging to include non-medical costs, we suggest to also include at least non-diagnostic costs in addition to the diagnostic costs. If studies work to conform to this approach, this would also improve insight in and appreciation of the cost-effectiveness of ES and GSCitation36. This is also supported by a study of Vrijenhoek et al.Citation29 which stated that benefit of genetic diagnostics can be broader than the diagnostic test itself. Having a diagnosis can also influence future health, treatment can be started earlier or expensive surgeries can be prevented.
Considering time horizon, no uniform method was used to determine the start and end of a study. Most of the studies ended the economic evaluation when a final diagnosis was found or when ES was initiated. However, the total duration included by the studies varied widely. This confirms the need for a uniform approach to ensure comparability. A study of Dragojlovic et al.Citation36 concluded that longer time horizons need to be included. Especially in patients without a conclusive diagnosis, long-term costs may influence conclusions about the cost-effectiveness of a diagnostic test. It is not possible to judge whether studies which took a fixed time-period as time horizon also took the complete diagnostic trajectory into account. To capture the full benefit of ES and/or GS, at least the complete diagnostic trajectory should be taken into account. According to Dragojlovic et al.Citation36, a longer follow-up period of at least 2 or 3 years is needed to capture the health benefit following having or not having a diagnosis. The latter may even suggest that stratification according to the outcome of the genetic test should be performed, as differences may be expected.
Interestingly, there seems to be a shift in focus on how effects were studied over time. The first publications regarding evaluations of NGS in pediatric genetics include effects directly related to the diagnostic trajectory, for instance diagnostic yield or cost-effectiveness itself. In health economic evaluations, a quality-adjusted life year (QALY) is most often used as an effect measure to calculate the incremental cost effectiveness ratio (ICER)Citation40. Since the end of 2017, it is common to use the ICER used to investigate cost-effectiveness within the field of health technology assessment. Although Stark et al.Citation27 did investigate QALYs, this was not included in an ICER. The first two studies which calculated an ICER included additional positive findings in scenario analysis as an effect measureCitation24,Citation25. Later on, effects were replaced by an additional number of diagnosesCitation26,Citation28. A more recent study of Schofield et al.Citation34 included QALYs to calculate the ICER. Although valuable, making decisions on willingness to pay per additional diagnoses is difficult. Besides, also no conclusive diagnosis can have added value to both doctors and (parents of the) patients, for example, in the case of a severe disease. Expensive care might be continued if no severe diagnosis can be found. For decision-makers, QALYs are very well-known parameters in the decision-making process. Other outcome measures, such as clinical utility, are more difficult to base reimbursement decisions upon, although these better reflect the added clinical value within the pediatric population. Findings of this study again demonstrated that more discussions should be initiated with all decision-makers to gain complete insight in most valuable other outcome measures compared to the QALY to base a decision upon.
Remarkably, two studies included the number of ongoing pregnancies and the utilization of parents’ reproductive genetic services as effect measuresCitation27,Citation34. This result also suggests that including costs goes beyond including costs directly related to the patient (child) him/herself. That is, in daily practice, guidance based on genetic test results is often not limited to impacting (future) life decisions of the patient him/herself, but also affects the choices and healthcare costs of his/her (blood) relatives. In order to perform economic evaluations in NGS, it is important to include all relevant information. However, at the moment it is unclear how this relevant information can be defined and to what extent it is needed to involve effect measures related to the relatives of the child. These findings confirm the need of a uniform approach, including all cost aspects, which should be the minimum requirement to guide decision-making.
Recently, it has been outlined that non-adherence to current health economic checklists is a major issue in economic evaluations of NGS for pediatric patientsCitation6. Although Alam and SchofieldCitation6 make a valid point regarding non-adherence, they did not discuss that disease-specific guidance to perform an economic evaluation in a certain setting was lacking. Examples include for instance that the use of terminology by the included papers was very diverse (), but also that is was unclear what was meant by the terms in the absence of definitions, making comparisons and interpretation difficult. Whereas these at large may depend on the study perspective, also these were not clearly outlined. The end results are economic evaluations within pediatrics for which it is underdefined which costs to include or how to define these. Such practical, disease-specific guidance is needed to ensure the improvement of future economic evaluations in genetics and thereby ensure the correct collection of highly valid data which can be used by the entire scientific field. These improvements can be created by more guidance as many authors of economic evaluations take previous papers as an example when deciding upon inclusion of costs; high quality standards should therefore be developed and also journals should be made aware of the importance of so-called cross-validation.
It was unclear whether or not studies had the intention to perform a full economic evaluation. In order to create more uniformity and comparable studies, it is essential that studies indicate whether the objective was to conduct a full-economic evaluation and/or whether there was a transparent reason to include certain costs and or effects in the evaluation.
The variability in choice of costs and effects also indicates the lack of discussion and guidance with current decision-makers within the field. It is highly relevant that such a discussion takes place to ensure inclusion of the most valuable outcome measures for making decisions on reimbursement and timing of expensive diagnostics. Within pediatric genetics, disease-specific guidance as outlined in this review is needed. This guidance would increase the quality, reliability, and comparability of outcomes. Current initiatives such as the GEECS (Global Economics and Evaluation of Clinical Genomics Sequencing Working Group) are essential in this caseCitation41. Within GEECS, for instance, they focus on improving methods used for assessing the value of new genomic technologies. The lack of uniformity and consensus on outcome measures (as indicated in this study) indicates that guidance and consensus on outcome measures should have high priority to ensure and improve decision-making.
Finally, the majority of the included evaluations in recent reviews have had a focus on the implementation of ESCitation13–20,Citation22,Citation23,Citation25,Citation27–35,Citation37,Citation38, and only six studies investigated the implementation of GSCitation12,Citation21,Citation24,Citation26,Citation36,Citation39. In order to help decision-makers with future decisions, especially in a fast-developing field such as medical genetics, it would be very relevant to outline which outcomes and individual input parameters are useful for future decisions on for instance implementation of GS in clinical practice. If achieved, this will also improve the power of evidence and better guide future research on cost-effectiveness studies towards those areas for which evidence is still largely unexplored. Within the field of medical genetics, much effort is being made to share knowledge on genetic causes of disease. Similar efforts should be made to push the field of economic analysis in genetics. Hereto, transparency in sharing outcomes and knowledge with regard to health economic evaluation is essential.
In general, it is challenging to use current health economic methods to capture the full benefit of NGS-related diagnostics. For instance, the inclusion of secondary findings, non-health benefits, and family spillover effects are difficult to incorporate in cost savings, let alone in one overarching number informing decision-makers. New approaches and methods should become available to fully capture, address, and evaluate the added benefit. Methodological research is essential in order to more precisely estimate the impact of genetic diagnostics and to improve well-informed/valid decisions in the near future.
This study has two (minor) limitations. First of all, only Pubmed/MEDLINE and EMBASE were searched. However, we also manually inspected the reference lists of the included studies to ensure that all relevant studies were included in this review. This did not lead to any new inclusions. Another possible limitation of this study was that we did not perform quality checks for the included studies. Although we did use the PRISMA reporting guidelinesCitation11, no alternative quality checks were performed. However, we do not think that inclusion of extra databases or adding quality checks would result in different results and/or conclusions.
Conclusions
In conclusion, a large variety in choice of costing characteristics and outcome measures is present in economic evaluations of new genetic technologies in pediatric conditions. This variability, shown by the included studies, indicates randomness in methods for economic evaluations in NGS and hampers a reputable comparison between outcomes. We argue that an improvement in cost collection (duration and type of cost data) and standardization in outcome measure is necessary for valid decision-making in the field of pediatric genetics and beyond. In addition to collaboration on clinical outcomes and data sharing, we should strive for a uniform approach in health economics in genetics to fight research waste but even more to speed up and improve the decision-making process; now is the time.
Transparency
Declaration of funding
This work was financially supported by grants from the Netherlands Organization for Health Research and Development (ZonMw) [843002608 and 846002003 to L.E.L.M. Vissers and J.K. Ploos van Amstel]. The aims of this study also contribute to the Solve-RD project (to L.E.L.M. Vissers) which has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 779257.
Declaration of financial/other interests
The authors have no conflict of interest to declare. Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
Acknowledgements
The results of this study contribute to the goals of the RADICON-NL consortium, which aims to increase diagnosis for patients with rare diseases in a shortened time frame, thereby reducing the complexity and overall costs associated with obtaining this diagnosis.
References
- Ayme S, Schmidtke J. Networking for rare diseases: a necessity for Europe. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2007;50(12):1477–1483.
- Zurynski Y, Frith K, Leonard H, et al. Rare childhood diseases: How should we respond? Arch Dis Child. 2008;93(12):1071–1074.
- Bick D, Dimmock D. Whole exome and whole genome sequencing. Curr Opin Pediatr. 2011;23(6):594–600.
- Vrijenhoek T, Kraaijeveld K, Elferink M, et al. Next-generation sequencing-based genome diagnostics across clinical genetics centers: Implementation choices and their effects. Eur J Hum Genet. 2015;23(9):1142–1150.
- Koboldt DC, Steinberg KM, Larson DE, et al. The next-generation sequencing revolution and its impact on genomics. Cell. 2013;155(1):27–38.
- Alam K, Schofield D. Economic evaluation of genomic sequencing in the paediatric population: a critical review. Eur J Hum Genet. 2018;26(9):1241–1247.
- Buchanan J, Wordsworth S, Schuh A. Issues surrounding the health economic evaluation of genomic technologies. Pharmacogenomics. 2013;14(15):1833–1847.
- National Institute for Health and Care Excellence (NICE) [Internet] Guide to the methods of technology appraisal 2013. [cited 2021 Feb 25]. Available from: https://www.nice.org.uk/process/pmg9/resources/guide-to-the-methods-of-technology-appraisal-2013-pdf-2007975843781.
- Schwarze K, Buchanan J, Taylor JC, et al. Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genet Med. 2018;20(10):1122–1130.
- Drummond MF, Sculpher MJ, Claxton K, et al. Methods of economic evaluation in healthcare programmes. 4th ed. Oxford: Oxford University Press; 2015.
- Moher D, Liberati A, Tetzlaff J, The PRISMA Group, et al. Preferred reporting items for systematic reviews and Meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097.
- Soden SE, Saunders CJ, Willig LK, et al. Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Sci Transl Med. 2014;6(265):265ra168.
- van Nimwegen KJ, Schieving JH, Willemsen MA, et al. The diagnostic pathway in complex paediatric neurology: a cost analysis. Eur J Paediatr Neurol. 2015;19(2):233–239.
- Valencia CA, Husami A, Holle J, et al. Clinical impact and cost-effectiveness of whole exome sequencing as a diagnostic tool: a pediatric center’s experience. Front Pediatr. 2015;3:67.
- Joshi C, Kolbe DL, Mansilla MA, et al. Reducing the cost of the diagnostic odyssey in early onset epileptic encephalopathies. Biomed Res Int. Epub. 2016;2016:1–8.
- Sabatini LM, Mathews C, Ptak D, et al. Genomic sequencing procedure microcosting analysis and health economic cost-impact analysis: a report of the association for molecular pathology. J Mol Diagn. 2016;18(3):319–328.
- Nolan D, Carlson M. Whole exome sequencing in pediatric neurology patients: Clinical implications and estimated cost analysis. J Child Neurol. 2016;31(7):887–894.
- Monroe GR, Frederix GW, Savelberg SM, et al. Effectiveness of whole-exome sequencing and costs of the traditional diagnostic trajectory in children with intellectual disability. Genet Med. 2016;18(9):949–956.
- Schofield D, Alam K, Douglas L, et al. Cost-effectiveness of massively parallel sequencing for diagnosis of paediatric muscle diseases. NPJ Genom Med. 2017;2:4.
- Vissers L, van Nimwegen KJM, Schieving JH, et al. A clinical utility study of exome sequencing versus conventional genetic testing in pediatric neurology. Genet Med. 2017;19(9):1055–1063.
- Hayeems RZ, Bhawra J, Tsiplova K, et al. Care and cost consequences of pediatric whole genome sequencing compared to chromosome microarray. Eur J Hum Genet. 2017;25(12):1303–1312.
- Stark Z, Schofield D, Alam K, et al. Prospective comparison of the cost-effectiveness of clinical whole-exome sequencing with that of usual care overwhelmingly supports early use and reimbursement. Genet Med. 2017;19(8):867–874.
- Tan TY, Dillon OJ, Stark Z, et al. Diagnostic impact and cost-effectiveness of whole-exome sequencing for ambulant children with suspected monogenic conditions. JAMA Pediatr. 2017;171(9):855–862.
- Tsiplova K, Zur RM, Marshall CR, et al. A microcosting and cost-consequence analysis of clinical genomic testing strategies in autism spectrum disorder. Genet Med. 2017;19(11):1268–1275.
- Dillon OJ, Lunke S, Stark Z, et al. Exome sequencing has higher diagnostic yield compared to simulated disease-specific panels in children with suspected monogenic disorders. Eur J Hum Genet. 2018;26(5):644–651.
- Yuen T, Carter MT, Szatmari P, et al. Cost-effectiveness of genome and exome sequencing in children diagnosed with autism spectrum disorder. Appl Health Econ Health Policy. 2018;16(4):481–493.
- Stark Z, Schofield D, Martyn M, et al. Does genomic sequencing early in the diagnostic trajectory make a difference? A follow-up study of clinical outcomes and cost-effectiveness. Genet Med. 2019;21(1):173–180. Corrected and republished from: Genet Med. 2019;21(2):516.
- Howell KB, Eggers S, Dalziel K, et al. A population-based cost-effectiveness study of early genetic testing in severe epilepsies of infancy. Epilepsia. 2018;59(6):1177–1187.
- Vrijenhoek T, Middelburg EM, Monroe GR, et al. Whole-exome sequencing in intellectual disability; cost before and after a diagnosis. Eur J Hum Genet. 2018;26(11):1566–1571.
- Stark Z, Lunke S, Brett GR, et al. Meeting the challenges of implementing rapid genomic testing in acute pediatric care. Genet Med. 2018;20(12):1554–1563.
- Palmer EE, Schofield D, Shrestha R, et al. Integrating exome sequencing into a diagnostic pathway for epileptic encephalopathy: evidence of clinical utility and cost effectiveness. Mol Genet Genomic Med. 2018;6(2):186–199.
- Demos M, Guella I, DeGuzman C, et al. Diagnostic yield and treatment impact of targeted exome sequencing in early-onset epilepsy. Front Neurol. 2019;10:434.
- Radio FC, Ruzzeddu M, Bartuli A, et al. Cost-effectiveness of exome sequencing: an Italian pilot study on undiagnosed patients. New Genet Soc. 2019;38(3):249–263.
- Schofield D, Rynehart L, Shresthra R, et al. Long-term economic impacts of exome sequencing for suspected monogenic disorders: diagnosis, management, and reproductive outcomes. Genet Med. 2019;21(11):2586–2593.
- Yokoi TA, Enomoto Y, Tsurusaki Y, et al. An efficient genetic test flow for multiple congenital anomalies and intellectual disability. Pediatr Int. 2020;62(5):556–561.
- Dragojlovic N, van Karnebeek CDM, Ghani A, et al. The cost trajectory of the diagnostic care pathway for children with suspected genetic disorders. Genet Med. 2020;22(2):292–300.
- Kosaki R, Kubota M, Uehara T, et al. Consecutive medical exome analysis at a tertiary center: diagnostic and health-economic outcomes. Am J Med Genet A. 2020;182(7):1601–1607.
- Smith HS, Swint JM, Lalani SR, et al. Exome sequencing compared with standard genetic tests for critically ill infants with suspected genetic conditions. Genet Med. 2020;22(8):1303–1310.
- Yeung A, Tan NB, Tan TY, et al. A cost-effectiveness analysis of genomic sequencing in a prospective versus historical cohort of complex pediatric patients. Genet Med. 2020;22(12):1986–1993.
- Drummond M, Brixner D, Gold M, et al. Toward a consensus on the QALY. Value Health. 2009;12(Suppl 1):S31–S35.
- Global Economics and Evaluation of Clinical Genomics Sequencing Working Group (GEECS) [Internet, cited 2021 Mar 15]. Available from: https://www.geecsecon.org/.