ABSTRACT
Background
Cell-free DNA analysis in cancer has gone from research to widespread clinical use in the past 10 years. At Memorial Sloan Kettering Cancer Center, we developed a technology and test to assay cell-free DNA (cfDNA) from blood (plasma) in our retinoblastoma patients.
Results
cfDNA derived from intraocular retinoblastoma can be measured and quantified in the blood (plasma) of patients. It is derived from the tumor cells themselves. Simulating lesions did not have cfDNA abnormalities. cfDNA disappears quickly after cutting the optic nerve (50% gone in 10 minutes) and if cfDNA is measurable after enucleation, metastases develop. Analysis of the buffy coat can detect germline defects including very low levels of mosaicism not detected with other NGS techniques. Analysis of the buffy coat also reveals non Rb1 germline predilections to second cancers.
Conclusion
Analysis of cfDNA from blood of retinoblastoma patients can be used to diagnose and manage retinoblastoma and reflect an accurate molecular profile of RB1 abnormalities of the intraocular tumor. Analysis of the germline with the buffy coat detects very low levels of mosaicism not detected with conventional methods. Liquid biopsy for retinoblastoma is already in clinical use and offers information not available with any other technique.
Introduction
There is great interest in exploring cell-free DNA (cfDNA) in cancer for diagnosis, differential diagnosis, prognosis, progression, choosing appropriate therapy, monitoring the development of new clones, and predicting outcome (Citation1). In 2011, there were fewer than 200 papers in the world literature on the subject and now, ten years later more than 4,000. Cell-free DNA has been identified in sputum, ascites, pleural effusions, CSF, stool, and urine in adult and pediatric cancers and in the aqueous of retinoblastoma patients. It has rapidly gone from a subject of “interest” to clinical use in oncology worldwide.
At Memorial Sloan Kettering Cancer Center, we created a team of scientists and clinicians with expertise in cfDNA and created a methodology for studying cfDNA in cancer with particular interest in retinoblastoma. This lecture summarizes what we know and what we do not know as of my Ellsworth Lecture which was delivered on 3 September 2021 before the International Society of Genetic Eye Diseases (ISGED). Though I was honored to deliver the lecture this work is the result of a collaborative effort and I want to acknowledge the important contributions of my colleagues: Diana Mandelker PhD, Rose Brannon PhD, Ryma Benayed PhD, Michael Berger PhD, Maia Arcila MD, Mark Ladanyi MD, Danielle Friedman MD, Gowtham Jayakumaram MS, Melissa Robbins MPH, Diana Haggag-Lindgren BS, Neeray Shulka MD, and Michael Walsh MD. The original work was performed by Prachi Kothari DO and Diana Tsui PhD and I especially grateful for the assistance of my retinoblastoma colleagues Ira Dunkel MD and Jasmine Francis MD.
Definitions
cfDNA
cfDNA refers to any DNA (usually double stranded) that is extracellular. It has been identified in sputum, ascites, pleural effusions, CSF, stool and urine, and aqueous humor. These are small fragments and have a relatively short half-life outside the cell (hours). The origin of cfDNA is thought to be primarily from necrotic and apoptotic cells, cfDNA crosses the placenta, and may represent as much as 10% of a mother’s circulating DNA in the first trimester.
ctDNA
ctDNA refers to extracellular DNA fragments specifically derived from cancer cells. These tend to be smaller than cfDNA, often having only 50–150 base pairs.
RB1
Refers to the retinoblastoma protein and RB1 refers to the gene. cfDNA of RB1 is ctDNA.
VAF
VAF refers to variant allele frequency and it is the way percentages of cf DNA in the blood are expressed. It is the proportion of allele bearing variants over the total number of wild plus variant alleles.
The MSK test
MSK created a test called “ACCESS” (Analysis of Circulating cfDNA to Evaluate Somatic Status) to identify and measure cfDNA (Citation2). To do this, they sequenced more than 25,000 solid tumors which gave a list of 129 genes of 826 exons that were most common in these cancers (). The test also interrogates 560 common SNPs and 40 introns known to be involved in cancer rearrangements. ACCESS utilizes Next Generation Sequencing (NGS) and is done on plasma (10–20cc) or other extracellular fluids with a turnaround time of 2–3 weeks. Originally a research project, in May 2019, it was approved by New York State as a clinical test and is now offered to all cancer patients at MSK. Details of the test have been previously published in detail (Citation2).
Table 1. MSK-ACCESS gene panel.
ACCESS uses matched white blood cells (WBCs) from the buffy coat of the same specimen which enables it to filter out more than 10,000 germline and clonal hematopoiesis variants. This is the reason for its sensitivity; it detects single nucleotide polymorphisms (SNPs) to 0.1%, Insertions or deletions (Indels) to 0.1% and copy number alterations (CNA) at a fold change of 1.5. Prior publications have demonstrated that it identifies molecular alterations in more than 80% of all solid cancers studied.
ACCESS tests for all exons of RB1 can detect missense, inframe, truncating, fusion, and other molecular aberrations of the gene.
MSK also developed a somewhat similar test (also NY State approved) that is done on tissue and it is called IMPACT (Integrated Mutation Profiling of Actionable Cancer Targets). It tests 529 cancer-related genes and RB1 is one of these genes too. Because the tissue results are compared to simultaneous blood drawn from the patient IMPACT, like ACCESS filters out germline and clonal hematopoiesis variants.
Is ctDNA present in blood of naïve retinoblastoma patients?
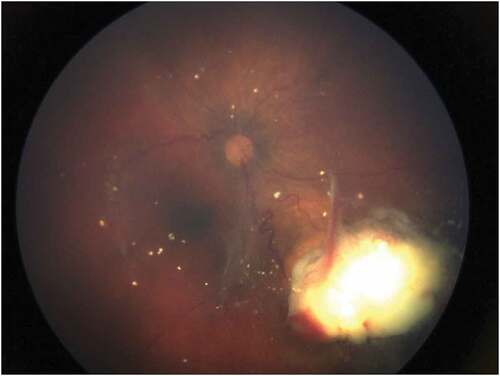
After 4 years of refining our technique, we tested the blood of 13 unilateral patients who had not received any treatment and detected ctDNA (RB1) in 10 of the patients (Citation3). By 2021, we reported that 18 of 21 tested samples were positive (Citation4). VAF levels were 1.63–12.6%. At the present time, we do not know why some of the specimens were negative. In cancer, in general, it is thought that tumors smaller than 10 mm will not be detectable in blood (Citation5), but some of our tumors were smaller than 10 mm and had measurable VAFs. represents a photograph of a recurrent tumor in one eye that was only 2–3 DD in size and still had ctDNA detected from blood.
Is the RB1 ctDNA in the blood coming from the tumor?
In 82 cases, we had molecular information about tissue from IMPACT (after enucleation) that we could correlate with plasma ACCESS levels. In every case, the abnormality was the same so yes, the plasma ctDNA accurately reflected the tissue DNA.
Do retinoblastoma simulating lesions have ctDNA?
We have now looked at 12 bloods from patients with Coats disease, 1 Persistent Fetal Vascular Syndrome, 1 medulloepithelioma, and 1 intraocular teratoma and no ctDNA was found. Assaying plasma ctDNA may be useful in the differential diagnosis of leukocoria and may help minimize clinical errors in the future.
Do initial VAFs predict subsequent metastases?
In our first two papers, we emphasized that naive patients with the highest levels of circulating ctDNA went on to develop metastases (Citation3,Citation5), but the numbers were small. In the US, only 4% of retinoblastoma patients develop metastases (Citation6). To date, all patients with initial VAF over 8.1% have gone on to develop metastases (3 patients) and no patient with a VAF below 3.2% has developed metastases (127 patients). Although we do have patients with VAFs between these cut off values, follow-up is short though to date none has developed a metastasis.
For these few patients, there appears to be a relationship between the initial VAF and the time from treatment to the development of clinical metastases. The higher the initial VAF the shorter time to develop metastases. For example, a patient with an initial VAF of 12.6% had metastases within 3 months, while one with an initial VAF of 8.1% took 6 months to develop metastases.
What happens to the plasma ctDNA after treatment?
After enucleation
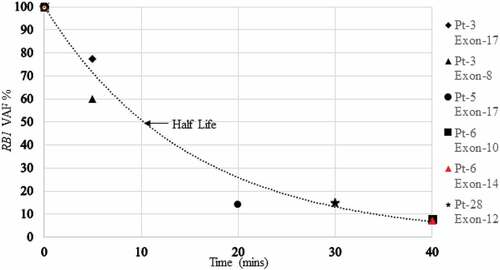
Ideally, we would draw blood prior to enucleation and then at multiple time periods during and after enucleation for each patient to chronicle the changes in VAF during and after enucleation. Because the test presently requires 10–20cc for each test that is impossible, so we collected blood before enucleation and then at different time periods for patients. Although the actual initial VAFs were similar, they were not identical, so we normalized the levels calling each initial pre enucleation VAF 100%. The graphic representation of this is presented below: .
The VAF decreased by 50% within 10 min of severing the optic nerve and 90% within 40 min (before the operation was completed).
Is ctDNA detectable in unilateral retinoblastoma patients after enucleation?
We have now tested 57 patients days, months, and years (up to 21 years) after enucleation for unilateral retinoblastoma. In 55 of the 57 patients, there was no ctDNA present in plasma and none of the 55 developed metastasis. The two exceptions are of interest.
One unilateral patient did not have blood drawn prior to enucleation but 5 days after the enucleation blood was drawn and the ctDNA VAF was 0.42%. Ten months later, metastatic disease developed.
A second unilateral patient also did not have blood drawn prior to enucleation but 10 months after enucleation ctDNA was drawn and was 94.5%. Work-up revealed metastatic disease (the child was asymptomatic).
Collectively, these results suggest that if there is any measurable ctDNA after enucleation for unilateral retinoblastoma that metastases will develop.
After intraarterial chemotherapy
We recently published on the fate of ctDNA after intraarterial chemotherapy (Citation7) in eyes where measurable ctDNA was present prior to treatment. Once again, we did not have complete sequential blood draws on each patient but we did have initial bloods on everyone and then at least one subsequent measurement from each patient. In 90% of cases, there was no measurable ctDNA when blood was studied one month later. One patient still had measurable VAF at 3 months. This suggests that ctDNA can be used as a guide to determine when intraarterial chemotherapy can be stopped (? after one treatment) and when it needs to be continued beyond the standard threetreatments.
VAF in untreated patients
Since we treat patients with retinoblastoma, it is unusual that we have sequential measurements in a patient who has not have treatment but in one case a patient had an initial VAF of 0.34%. For social reasons, no treatment was given for a month and one month later the VAF was 2.28%.
Taken together these results suggest that VAF can be used to guide treatment and that increasing levels of VAF suggest continued clinical activity.
Opaque media
Clinical activity in retinoblastoma is usually assessed with the indirect ophthalmoscope. Ultrasound and MRI can be helpful, but they are replete with false positives and negatives; assessing clinical activity (progression or regression of disease) in the face of opaque media is a significant challenge. We have had the opportunity to measure VAF in 13 treated eyes that developed vitreous hemorrhage, cataract, retinal detachment, or phthisis. In 13 cases, the VAF was negative and to date not one of these patients has developed orbital or metastatic disease. While clinicians are always cautious about making decisions based on negative tests (because of the possibility of false negatives), VAFs in opaque media eyes may add an additional test that may be useful in the overall assessment of clinical activity.
The use of ctDNA as screening for second non ocular tumors
Patients with the genetic form of retinoblastoma are at high risk for the development of subsequent “second” non ocular cancers (sarcomas, melanoma etc.) (Citation8–10). The rate is between ½ and 1% a year. In fact, more patients in the U.S. die from these second cancers than retinoblastoma itself. To date screening has been performed with imaging techniques but every study has shown that these strategies do not improve survival (Citation11). Something better is needed. MSK ACCESS interrogates 128 critical cancer genes in addition to RB1. To date, we have looked at the blood of 86 bilateral survivors and found three with somatic alterations in non-Rb cancer related genes. The first patient had FBXW7, the second patient had ALK, BRAF, KRAS APC, ATM, and CDH1. The third patient had BRCA1. These patients are asymptomatic and presently being worked-up for a silent malignancy.
What have we learned by analysis of the buffy coat?
As mentioned previously, ACCESS performs NGS on the buffy coat in addition to the plasma. This is used to filter out germline abnormalities and clonal hematopoiesis, but it can also reveal information about other germline abnormalities in the patient. Here is what we have learned to date:
ACCESS detects lower level mosaicism than presently available with other techniques. Many of our patients had genetic testing years ago with Sanger sequencing which could detect mosaicism at a level of about 20%. Modern NGS testing detects “low level mosaicism” (defined as less than 20%) and usually quoted as 5%. As ACCESS compares the somatic abnormality to the buffy coat, it detects mosaicism below 1%. We now have four patients who have had modern NGS commercial genetic testing on blood where no RB1 abnormality was detected but with ACCESS we found very low levels of mosaicism (less than 5%). We believe that very low levels of mosaicism are much more common (perhaps universal) than presently reported with conventional genetic testing.
ACCESS can detect non RB1 germline predilection to cancers. We recently published that 17% of all children at MSK with cancer harbor a germline predilection to cancer unrelated to the cancer for which they are being treated (Citation12). To date, 10% of all our bilateral retinoblastoma children have been found to have other germline mutations predisposing them to cancer. Among the genes identified are MSK3, MITF, Rad516-1, CHEK2 and NF1.
In summary, RB1 cell-free DNA was identified and quantified in most naive retinoblastoma patients, to date we have had no false positives as leukocoria look alikes have been negative, the highest VAF at diagnosis suggests that metastases will develop and possibly the higher the level the shorter the interval till metastases. Circulating RB1 cfDNA disappears quickly after enucleation and intraarterial chemotherapy. Persistence of circulating ctDNA after enucleation in unilateral retinoblastoma augurs metastases and patients with metastatic disease have high levels of circulating ctDNA. cfDNA may be useful in differential diagnosis of retinoblastoma and opaque media cases. ctDNA may be used to guide treatment decisions. cfDNA reveals the precise genetic abnormality of the intraocular tumor itself. These exciting findings need to be verified in more cases but we feel that they can serve as the basis for additional investigations by us and other retinoblastoma centers.
ACCESS analysis of the buffy coat reveals mosaicism that is not detected with other techniques and may also reveal additional, non-RB1 germline gene abnormalities that predispose children to cancers throughout their lives.
The dream of a blood test for cancer is here.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Corcoran RB, Chabner BA. Application of Cell-free DNA analysis to cancer treatment. N Engl J Med. 2018;379(18):1754–65. doi:10.1056/NEJMra1706174.
- Brannon AR, Jayakumaran G, Diosdado M, Patel J, Razumova A, Hu Y, Meng F, Haque M, Sadowska J, et al. Enhanced specificity of high sensitivity somatic variant profiling in cell-free DNA via paired normal sequencing using MSK-ACCESS. Nat Commun. 2021;12(3770). https://doi.org/10.1038/s41467-021-24109-5
- Kothari P, Marass F, Yang JL, Stewart CM, Stephens D, Patel J, Hasan M, Jing X, Meng F, Enriquez J, et al. Cell-free DNA profiling in retinoblastoma patients with advanced intraocular disease: an MSKCC experience. Cancer Med. 2020;9(17):6093–101. doi:10.1002/cam4.3144.
- Abramson DH, Mandelker D, Francis JH, Dunkel IJ, Brannon AR, Benayed R, Berger MF, Arcila ME, Ladanyi M, Friedman DN, et al. Retrospective evaluation of somatic alterations in Cell-Free DNA from blood in retinoblastoma. Ophthalmol Sci. 2021;1(1):100015. doi:10.1016/j.xops.2021.100015.
- Fiala C, Diamandis EP. Cell-free DNA analysis in cancer. N Engl J Med. 2019;380(5):501.
- Lu JE, Francis JH, Dunkel IJ, Shields CL, Yu MD, Berry JL, Kogachi K, Skalet AH, Miller AK, Santapuram PR, et al. Metastases and death rates after primary enucleation of unilateral retinoblastoma in the USA 2007-2017. Br J Ophthalmol. 2019;103(9):1272–77. doi:10.1136/bjophthalmol-2018-312915.
- Francis JH, Gobin YP, Brannon AR, Swartzwelder CE, Berger MF, Mandelker DL, Walsh MF, Dunkel IJ, Abramson DH. RB1 circulating tumor DNA in the blood of patients with unilateral retinoblastoma: before and after intra-arterial chemotherapy. Ophthalmol Sci. 2021;1(3). doi:10.1016/j.xops.2021.100042.
- Abramson DH, Ellsworth RM, Zimmerman LE. Nonocular cancer in retinoblastoma survivors. Trans Sect Ophthalmol Am Acad Ophthalmol Otolaryngol. 1976;81(3 Pt 1):454–57.
- Eng C, Li FP, Abramson DH, Ellsworth RM, Wong FL, Goldman MB, Seddon J, Tarbell N, Boice JD. Mortality from second tumors among long-term survivors of retinoblastoma. J Natl Cancer Inst. 1993;85(14):1121–28. doi:10.1093/jnci/85.14.1121.
- Abramson DH. Second nonocular cancers in retinoblastoma: a unified hypothesis the franceschetti lecture. Ophthalmic Genet. 1999;20(3):193–204. doi:10.1076/opge.20.3.193.2284.
- Friedman DN, Hsu M, Moskowitz CS, Francis JH, Lis E, Fleischut MH, Oeffinger KC, Walsh M, Tonorezos ES, Sklar CA, et al. Whole-body magnetic resonance imaging as surveillance for subsequent malignancies in preadolescent, adolescent, and young adult survivors of germline retinoblastoma: an update. Pediatr Blood Cancer. 2020;67(7):e28389. doi:10.1002/pbc.28389.
- Fiala EM, Jayakumaran G, Mauguen A, Kennedy JA, Bouvier N, Kemel Y, Fleischut MH, Maio A, Salo-Mullen EE, Sheehan M, et al. Prospective pan-cancer germline testing using MSK-IMPACT informs clinical translation in 751 patients with pediatric solid tumors. Nat Cancer. 2021;2:357–65.