Abstract
Context
MicroRNA (miRNA) is an important regulator of gene expression. Methamphetamine (METH) induces a variety of alterations in different systems by affecting gene expression, but the effects of METH on miRNA profiles need to be elucidated.
Objectives
This study develops a rat model of METH addiction, and analyzes the expression profile alterations of miRNA in nucleus accumbens (NAc) of the METH-addicted rats.
Materials and methods
Sprague-Dawley rats were administered 10 mg/kg METH or vehicle twice a day for 4 weeks. The addictive behaviour of rats was estimated by CPP test. The pathological changes of brain tissues were then observed by HE and Glee silver staining. The miRNA profile analysis of the NAc of the rats was performed using an Illumina HiSeq™ 2500 sequencing system.
Results
CPP test indicated that METH significantly prolonged the residence time of the rats in the drug box (from 307 ± 97 to 592 ± 96 s). The pathological staining showed the distorted axons, and fewer polarized neurons in the METH-treated rats. We further identified 40 differential miRNAs (17 up- and 23 down-regulated) and three novel miRNAs (novel 237, 296 and 501) that responded to METH. The bioinformatic analysis for the potential targets of the differential miRNA suggests that the downstream were concentrated in the Wnt signalling pathway, tuberculosis, toxoplasmosis, spliceosome, lysosome, and axon guidance.
Discussion and conclusions
A number of miRNAs responding to METH were identified in the NAc of rats. These METH-regulated miRNAs provide a new perspective for revealing the molecular mechanisms of METH addiction.
Introduction
In the past decades, drug addiction has been considered a chronic debilitating cerebral disease (Leshner and Koob Citation1999), which is one of the costliest diseases in current society. Methamphetamine (METH) is a highly addictive psychostimulant that is easily synthesized with relatively low-cost materials, making the drug inexpensive and readily available. As a result, this drug has become more and more popular in recent years, and its abuse is a growing issue all over the world. The euphoric effect of METH lasts for 4–24 h (Cook et al. Citation1993), and its extra methyl group makes it more potent than amphetamine and dextroamphetamine (Fasciano et al. Citation1997). In addition, drug tolerance rapidly results in increased drug consumption to get the same euphoric effects (Brauer and de Wit Citation1996). The National Institute on Drug Abuse (Chandler et al. Citation2016) reported that the abuse of METH in 2013 increased to 242,000 cases, and has become a major illicit drug problem in the United States. Similar concerns exist in countries such as the UK, Finland and Australia. There are 1.35 million people abusing METH among the 2.5 million drug addicts in China, accounting for 56.1% of all drug addicts. METH has replaced heroin as the most abused drug in China. Although several behavioural therapies, including cognitive behavioural therapy and emergency management interventions, have been clinically applied, there is no specific treatment for METH addiction (Hamamoto and Rhodus Citation2009). Therefore, the development of effective treatment methods has become a top priority.
METH addiction is a chronic and relapsing disease that can induce certain types of mental and cognitive disorders. A clinical study has shown that long-term use of METH exhibits defects in several cognitive areas related to learning and memory (Reichel et al. Citation2011). Animal studies reported that animals injected with METH showed stronger drug-seeking behaviours and impaired ability to recognize new objects (Rogers et al. Citation2008). In general, most drugs of abuse have a common mode of action in the cerebral pathway of the mesolimbic dopaminergic system, which is linked to motivation and reward by releasing the neurotransmitter dopamine (DA) (Di Chiara Citation2002). Alterations in dopamine signalling can result in neurological adaptation, thereby reducing the individual’s sensitivity to the drug and/or increasing the individual’s propensity to drug exposure. Previous studies have found that consumption of DA and serotonin in the nucleus accumbens (NAc) after treatment with neurotoxic METH can reduce spontaneous movement and increase METH-induced stereotypic behaviours (Wallace et al. Citation1999). METH-induced dopamine depletion by repeated administration may be responsible for a decrease in dopamine-stimulated release and an augment in the stereotypic responses. The NAc is one of the most important parts of the brain involved in the regulation of the reward pathway, and changes in gene expression caused by addictive drugs may lead to addictive behaviour. In addition, some studies have shown that dopamine-independent enhancement occurs in the NAc, suggesting that multiple inputs are involved in the activation of key enhancement circuits in this area (Koob Citation1992).
MicroRNAs (miRNAs) are a class of small non-coding transcripts involved in the fine-tuning of gene expression during brain differentiation and development (Kosik Citation2006). They are a large number of post-transcriptional gene regulatory molecules regulating the expression of synaptic proteins (Schratt Citation2009). The miRNAs in mature neurons show a high degree of diversity, with approximately 100 miRNAs present in developed neurons (Fiore and Schratt Citation2007). The high expression of certain miRNAs in the brain suggests that they may be involved in neuronal morphogenesis, memory formation, and drug addiction (Ashraf et al. Citation2006). Recent studies have further demonstrated that abnormal expression of some miRNAs may lead to synaptic dysfunction and contribute to psychological and neurological diseases (Beveridge et al. Citation2008). Evidence for the role of miRNAs in brain reward circuits is provided in a study that suggests that miRNAs are involved in the addiction to cocaine (Chandrasekar and Dreyer Citation2009), alcohol (Pietrzykowski et al. Citation2008), nicotine (Huang and Li Citation2009), METH (Bosch et al. Citation2015) and several other classes of drugs (He et al. Citation2010). Research on cocaine addiction has confirmed up-regulation of miR-181a and down-regulation of miR-124a in the addiction-related areas such as the NAc, ventral cap, hippocampus, and prefrontal cortex (Chandrasekar and Dreyer Citation2009).
Manipulation of changes in miRNAs in the NAc appears to be sufficient to attenuate or enhance drug-seeking behaviour (Chandrasekar and Dreyer Citation2011). It is generally believed that a single miRNA induced by drug abuse may alter the expression levels of many downstream target genes that regulate the neural mechanisms related to addiction (Saba et al. Citation2012). In mammalian cells, there are still many miRNAs to be detected, and of course little is known about the level or pattern of miRNA expression in drug addiction. Therefore, this study demonstrates the relationship between the use of METH and addictive behaviour in rats. This study also performs a full-spectrum miRNA analysis of METH addiction in rat NAc tissues to identify miRNAs associated with METH abuse and dependence. This study provides insights into the investigation of miRNAs which may be involved in the use of METH and the regulation of addiction. Therefore, the knowledge obtained could be used to design a drug treatment strategy.
Materials and methods
Animals and administration
Twenty male Sprague-Dawley rats aged 6 weeks and weighing 180–220 g were obtained from Kunming Medical University. The rats were randomly divided into an MA4 (methamphetamine treatment for 4 weeks) group and a control group of 10 rats each. All rats were kept in an air-conditioned room at a constant temperature of 25 °C. All experiments were done at the animal experimentation centre of the First Affiliated Hospital of Kunming Medical University. They were given free access to water and forage, and were fed for 7 days to help them adapt to the environment. Then, each of the MA4 rats was injected with METH intraperitoneally twice a day (at 8 AM and 6 PM) at a dose of 10 mg/kg. The rats from the MA4 group were injected for 4 weeks. The control group was injected with the same amount of normal saline. One week after stopping the drug, 10% chloral hydrate (3 mL/kg) was injected intraperitoneally. Under anaesthesia, the rats were decapitated and craniotomy was performed on ice. Separate striatum, NAc and hippocampus were isolated. One side of each was stored in a freezer at −80 °C and the other side was fixed with 10% formaldehyde solution (catalog No. C1040410015, Nanjing Chemical Reagent, Nanjing, China), conventional dehydration, and paraffin embedding. Experiments were conducted using protocols in accordance with the institutional ethical guidelines concerning the care and use of laboratory animals. All animal procedures were approved by the animal ethics committee of the First Affiliated Hospital of Kunming Medical University (approval code: 20180528).
Methamphetamine conditioned place preference (CPP)
Each of the above rats was subjected to CPP tests at the 7th, 14th and 28th days after treatment. A conditional device (TOP-Bright, Shanghai, China), consisting of a white box, a black box at both ends, and a small box in the middle, was used to determine the CPP of rats. The walls of black and white boxes were smooth, while the floor was rough in the black box and smooth in the white box. The black box had a metal rod floor, and pine bedding beneath the floor. The white box had white walls and a mesh floor with Plexiglas directly under the mesh, with white painted wood on the sides. The middle box is the passage connecting the black and white boxes. The apparatus was put in a laboratory room separate from the colony room with a white noise generator and audio speaker (ambient background of 70 dB). A video camera used to record the experimental sessions was suspended from the ceiling above the apparatus.
Before injecting METH, the rats were permitted to move freely between the two boxes to adapt for 3 days (2 times/day, 15 min each time) and the average residence time of each rat in white and black boxes was recorded.
In the post-conditioning phase, the CPP test of each group was performed after the last administration of methamphetamine for 24 h, and the measurement was the same as the baseline measurement. The time the rats spent in the white compartment was recorded within 15 min. The whole process was captured with a digital video camera (Nikon, Tokyo, Japan) to record the track of the rats.
HE staining
Paraffin tissue sections were dehydrated conventionally, including xylene used 3 times, anhydrous alcohol, 95% alcohol, 80% alcohol, 75% alcohol and distilled water successively. Paraffin tissue sections were stained with haematoxylin for 5 min, rinsed with tap water and blotted dry. Then, they were differentiated with the ethanolic acid for 30 s, soaked in warm water at 50 °C for 5 min, and drained. Eosin was used to incubate the samples for 2 min and routine dehydration, using 95% alcohol, anhydrous alcohol, xylene carbolic acid, and xylene in order. Finally, they were sealed with a neutral resin.
Glee silver staining
Glee silver solution was prepared as follows: 30 mL of 20% silver nitrate (catalog No. 209139, Sigma-Aldrich, St. Louis, MO, USA) solution was mixed with 20 mL of 95% alcohol, and concentrated ammonia was added drop by drop until the resulting precipitate just dissolved. Subsequently, 3 or 4 drops of ammonia water were added.
The brain tissue was fixed in 4% neutral formaldehyde buffer for 24 h at room temperature and dehydrated with 70% alcohol, 80% alcohol, 90% alcohol, 95% alcohol and absolute alcohol in sequence. Then, brain tissues were embedded in paraffin and serially sliced at a thickness of 4 μm. They were dewaxed sequentially with xylene, absolute ethanol, 90% alcohol, 80% alcohol, 70% alcohol and distilled water and washed 3 times with distilled water. Paraffin sections were dipped in 20% silver nitrate solution for 30 min at 37 °C and washed with 10% formalin for 10–15 s. Furthermore, paraffin sections were incubated with Glee silver for 30 s and washed with 10% formalin for 1 min. Paraffin sections were rinsed once with distilled water and incubated with 5% sodium sulphite for 1–2 min. Later, they were washed with tap water and dehydrated with 70% alcohol, 80% alcohol, 90% alcohol, 95% alcohol and anhydrous alcohol in order. Finally, paraffin sections were observed under the microscope and captured.
Immunofluorescence
The brain tissue was embedded, sectioned and dewaxed. Paraffin sections were fixed with immunostaining fixative (catalog No. P0098, Beyotime, Shanghai, China), and then washed twice with immunostaining washing solution (catalog No. P0106, Beyotime) for 5 min each time. Paraffin sections were then blocked with immunostaining blocking solution (Beyotime, P0102) for 1 h at room temperature. The primary antibody was diluted to the appropriate concentration with immunostaining primary antibody dilution (catalog No. P0103, Beyotime), which was used to incubate paraffin sections at 4 °C overnight. The primary antibody of NSE (neuron specific enolase, catalog No. AF5169) and GFAP (glial fibrillary acidic protein, catalog No. AF2594) was obtained from the R&D system (Minneapolis, MN, USA). Then, the sections were rinsed 3 times with washing solution for 5 min each time. Next, the fluorescently labelled secondary antibody was diluted with immunofluorescent secondary antibody dilution buffer (catalog No. P0108, Beyotime), and was used to incubate. After that, paraffin sections were washed 5 times with the washing solution for 10 min each time. The sections were observed with a fluorescence microscope and photographed.
RNA preparation and miRNA sequencing
The RNA extraction was carried out as previously described (Campos et al. Citation2020). In brief, 50–100 mg of tissue was placed in a 1.5 mL EP tube (RNase free), and 1 mL of Trizol (Thermo Fisher Scientific, Waltham, MA, USA) was added to homogenize. After that, tissue was let sit for 5 min at room temperature (RT). Chloroform (0.2 mL) was added into the EP tube, mixed vigorously for 30 s, and let sit for another 5 min at RT. Then, tissue was centrifuged for 10 min at 4 °C 12,000 g, and the liquid in the centrifuge tube was layered after centrifuging. The supernatant was carefully pipetted into a new 1.5 mL EP with a small pipette. Isopropanol (0.5 mL) was added, mixed vigorously for 30 s, and let sit for another 10 min at RT. Then, the tissue was centrifuged again for 10 min at 4 °C 12,000 g, leaving a precipitate at the bottom of the tube. 75% ethanol (1 mL, precooled at 4 °C) was added and the precipitate was suspended again. After that, tissue was centrifuged for 15 min at 4 °C 12,000 g and the supernatant was discarded. An appropriate amount of diethyl pyrocarbonate (DEPC) H2O was added at 65 °C to promote dissolution according to the amount of precipitation when the EP tube was airing. The tube was oscillated to dissolve the precipitate. Then, the concentration of total RNA was measured by Nanodrop™ (Thermo Fisher Scientific). The RNA samples were used for miRNA sequencing and analysis with an Illumina (San Diego, CA, USA) HiSeq™ 2500 sequencing system when RNA was qualified.
Western blotting
The tissues were fully lysed and sonicated, and the total protein was extracted. Then, the protein was quantified using a BCA Protein Assay Kit (Thermo Fisher Scientific). Polyacrylamide gel electrophoresis was performed using 10 μg protein from each sample. Then, the protein on the gel was transferred to the polyvinylidene fluoride (PVDF) membrane using semi-dry transfer, and the PVDF membrane was sealed with 5% skim milk powder for 2 h at room temperature. Thereafter, the membrane was probed with the primary antibody at 1:1000 dilution with phosphate buffer saline (PBS) against RNA-binding protein 8a (Rbm8a, catalog No. sc-32312, Santa Cruz Biotechnology, Santa Cruz, CA, USA), synaptotagmin-7 (SYT7, catalog No. sc-293343, Santa Cruz Biotechnology), 5-hydroxytryptamine receptor 1B (5-HTR1B, catalog No. A-AW5541, Abgent, San Diego, CA, USA), reticulon-4 (RTN4, catalog No. 13401, Cell Signalling Technology, Danvers, MA, USA), or synaptic vesicle glycoprotein 2 A (SV2A, catalog No. 66724, Cell Signalling Technology) at 4 °C overnight. The PVDF membrane was then washed three times with PBS-Tween for 8 min each. The secondary antibody was diluted with PBS, and was used to incubate for 1 h at room temperature, and the PVDF membrane was washed again as before. β-actin was used as a loading control. Proteins were visualized using an enhanced chemiluminescence solution.
Quantitative real-time PCR
Total RNA was reverse transcribed to cDNA by using a Reverse Transcription Kit (Takara, Dalian, China). Real-time PCR analyses were performed with SYBR Green (Takara, Dalian, China) and followed the amplification parameters at 95 °C for 1 min, 95 °C for 10 s, and 60 °C for 60 s (40 cycles). The amplification was in the mixture of 50 ng cDNA, 2 mM Mg2+, 100 pmol primers of each, 200 mM dNTP, and 2.5 U Tag DNA polymerase. Results were normalized to the expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and the relative expression of genes was calculated using the 2−△△Ct method. The primers used were validated in other studies, and are listed as follows: Rbm8a, F: 5′-GCGTGAGGATTATGACAGCGTG-3′, R: 5′-TTCGGTGGCTTCCTCATGGACT-3′ (Liang et al. Citation2017); SYT7, F: 5′-ACTCCATCATCGTGAACATCATC-3′, R, 5′-TCGAAGGCGAAGGACTCATTG-3′ (Wu et al. Citation2020); Htr1b, F: 5′-CACCAACCTCTCCCACAACT-3′, R: 5′-CCAGAGAGGCGATCAGGTAG-3′ (Zajac et al. Citation2018); Rtn4, F: 5′-TCCTCGGGCTCAGTGGTTGTTG-3′, R: 5′-TGCCCTGAATGGGTGGCCTT-3′ (Rodríguez-Feo et al. Citation2015); Sv2A, F: 5′-TCTCTGCTCCAGGTGTTCCA-3′, R: 5′-CGGAGGCTAAGGTTTATTGCTAC-3′ (Shi et al. Citation2015); GAPDH, F: 5′-AGGTGAAGGTCGGAGTCA-3′, R: 5′-GGTCATTGATGGCAACAA-3′. The ABI 7500 fast real-time PCR system (Thermo Fisher Scientific) was employed for the PCR analyses.
In the quantitative PCR for miRNAs, they were ligated to 3′ adaptors and reverse transcribed to cDNAs with the parameters of 37 °C for 60 min and then 85 °C for 5 min. The primers for each miRNA were listed in Supplementary Table S1.
Statistical analysis
Statistical analysis was performed using PASW Statistics for Windows, Version 18.0 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism Software (GraphPad Software, Inc., San Diego, CA, USA). All comparisons were performed using Student’s t-test. p < 0.05 was considered to indicate a significant difference. Each experiment was performed at least three times, and the results were expressed as the mean ± standard deviation (SD).
Results
The effects of METH on behavioural changes, neurons, and axons
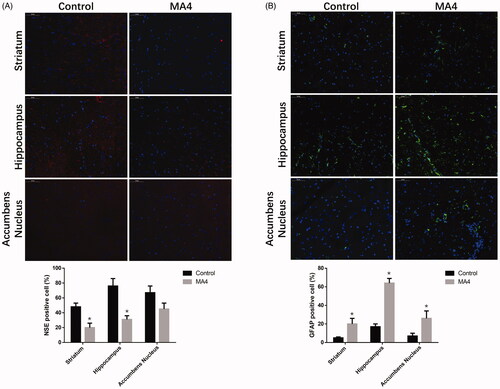
After training, the residence time of the METH-treated rats at the drug box side was gradually prolonged compared with the control rats. During the treatment, the time was increased from 307 ± 97 to 592 ± 96 s (p < 0.05) (). Glees silver staining showed that the striatum axons were more distorted, serpentine, and showed segmental thickening. The axonal gap was wider in the rats from the MA4 group compared with the control (). In the HE staining results, the neurons in the hippocampus of the control rats were more elliptical or conical with a large nucleoplasm ratio, and they had light blue-violet nuclei and clear nucleoli. In the MA4 group, there was a large number of neurons stained with cytoplasm; the cells were disordered and had unclear nuclei. The neurons in the NAc and striatum of the MA4 group rats were more rounded and less polarized. The Nissl body was also decreased. In addition, we observed the glial over proliferated in the striatum and hippocampus, and more glial nodules in the striatum caused by the METH treatment ().
Figure 1. The effects of METH on neurons and axons. (A) Glees silver staining revealed the morphological changes of axons in the striatum. (B) HE staining showed the pathological changes in the striatum, hippocampus, and nucleus accumbens.
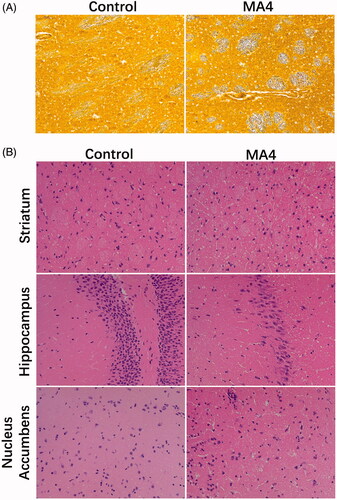
Table 1. The residence time in CPP test.
The effects of METH on neuron differentiation
Next, we observed the differences in neuron differentiation between the control group and METH-treated rats. The immunofluorescence indicated that NSE expressing cells in the striatum and hippocampus of the MA4 rats were scattered, and the number of NSE-positive cells decreased in the MA4 group compared with that of the control (). However, the density of GFAP positive astrocytes in the striatum and hippocampus was significantly increased in the MA4 group ().
Global miRNA sequencing analysis
A total of 40 differentially transcribed miRNAs (p value < 0.01 & |log2 (fold change)| > 1 in comparison with the control group (), 17 up-regulated and 23 down-regulated by METH treatment, were found with quantitative information and included in the next bioinformatic analyses (). Differential miRNAs were verified using qPCR, and their relative abundance was consistent with the results of the sequencing analysis (). The potential targets of the changed miRNAs were annotated according to their biological process, cellular component, and molecular function by BLAST2TO (). Biological processes analysis showed that these proteins were mainly involved in cellular component regulation, localization regulation, regulation of transport, intracellular signal transduction, and organic substance transport. We observed that most of the targets of the differential miRNAs were located in the intracellular region and organelles in the cellular component analysis. Molecular function analysis revealed that a large proportion of the targets played a role in protein binding, enzyme binding, kinase binding, and transferase activity.
Figure 3. Screening of differentially expressed miRNAs in rat NAc after METH treatment. (A) The volcano map of the differential miRNAs. The abscissa represents the fold change of miRNA expression, and the ordinate represents the statistical significance of the change in miRNA expression. The scatter points represent individual miRNAs. (B) Differential miRNA clustering map.
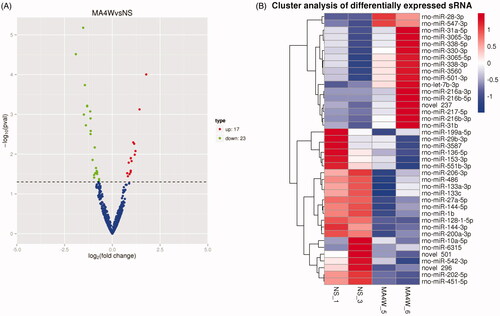
Figure 4. Validation of the differential miRNAs by qPCR. The top up-regulated and downregulated miRNAs in sequencing were measured in the NAc samples with or without METH treatment. Compared with the NC group, *p < 0.05.
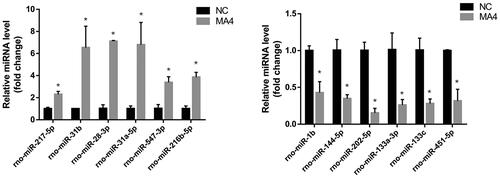
Figure 5. Enrichment analysis of target genes of differential miRNA. (A) GO enrichment analysis of the target genes of differentially expressed miRNAs. The three categories from left to right are biological processes, cellular components and molecular functions. (B) KEGG enrichment analysis of candidate target genes. The ordinate represents the pathway name, and the abscissa represents the rich factor. The size of the points indicates the number of the genes in the pathway.
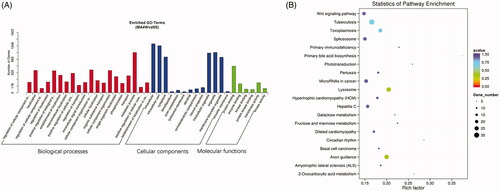
Pathway enrichment analysis for the targets of the changed miRNAs by KEGG demonstrated that these molecules are mainly involved in the Wnt signalling pathway, tuberculosis, toxoplasmosis, spliceosome, lysosome, axon guidance, etc. (). The above pathways might be impacted by the METH treatment and involved in the mechanisms of METH addiction.
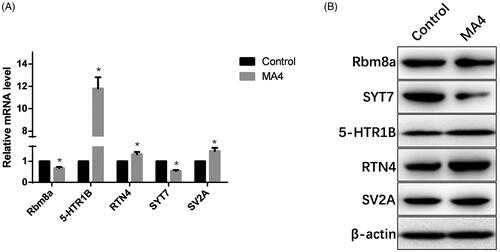
Validation of the potential miRNA target genes with qPCR
We further detected the expression levels of the several predicted target genes of the changed miRNAs. According to the results of qPCR, 5-HTR1B dramatically increased, and RTN4 and SV2A slightly increased as a result of METH treatment. Rbm8a and SYT7 decreased at the transcription level in MA4 rats (). Meanwhile, the protein level of these molecules was visualized by western blotting, which was in line with what was observed at the mRNA level ().
Discussion
The alterations of miRNA expression profiles caused by METH addiction are largely unclear. The current study documented the comparative miRNA profiles in the NAc of control and chronically METH-treated rats by sequencing. We identified a number of miRNAs that responded to METH. These METH-regulated miRNAs provide new perspectives for revealing the molecular mechanisms of METH addiction.
METH is a powerful psychological stimulator which has multiple effects on the nervous system. Besides its effects on neuronal plasticity and drug addiction, chronic METH consumption also induces autophagy in neurons, leading to degeneration of dopaminergic neurons (Larsen et al. Citation2002). Moreover, recent studies have shown that prolonged exposure to METH can lead to metabolic dysfunction, including disorders of amino acid and lipid metabolism (Bu et al. Citation2013). In addition, METH at both low and high doses can cause a heightened inflammatory environment in the brain (Nakajima et al. Citation2004), and thereby affect the regulation of cellular immune responses. In the present study, the potential target genes of the differential miRNAs regulated by METH are significantly enriched in pathways associated with Wnt signalling, tuberculosis, toxoplasmosis, spliceosome, lysosome, and axon guidance, suggesting that METH impacts these biological events by modulating the related miRNAs.
Previous studies have revealed that miRNAs play an important role in neuronal plasticity and drug addiction (Schratt Citation2009). The expression of miR-29b and miR-138 was found to be associated with neuronal plasticity (Siegel et al. Citation2009), and another study reported that miR-124 regulated cocaine-induced conditioned place preference (Chandrasekar and Dreyer Citation2011). We could deduce from the results that the differential miRNAs in the NAc may promote METH addiction by modulating the rewarding effect of METH and neuronal plasticity. Furthermore, expression of miR-29b in the NAc was downregulated by cocaine (Chandrasekar and Dreyer Citation2009), consistent with the trend in the current study. Similar changes of miR-29b in different data sets using different drugs and different treatment programmes were observed in the NAc, which may reflect the general role of the miRNA in regulating drug addiction.
RNA High-throughput sequencing is a very powerful tool for identifying new transcripts (Metzker Citation2010). We herein observed three new miRNA candidates (novel 237, 296 and 501) that responded to METH, but additional experiments are needed to determine their authenticity.
MiRNAs are capable of inhibiting gene expression at the mRNA level (Siomi and Siomi Citation2009). Therefore, we examined the potential targets of the differential miRNAs. Unexpectedly, the transcriptional data revealed that some potential target genes did not exhibit opposite changes to their associated miRNAs. This may be caused by the fact that mRNAs are not only regulated by miRNAs. Another possibility is that METH directly alters mRNA expression. We found in the pathway enrichment analysis that lysosome related genes were at the top rank. A recent study indicated that the METH treatment upregulated α-syn in neurons, which may directly induce mitochondrial damage, myelin sheath destruction, and synaptic failure. The excess α-syn might also indirectly promote tau phosphorylation through tau kinase, glycogen synthase kinase 3 beta (GSK3β) and cyclin-dependent kinase 5 (CDK5), leading to microtubule depolymerization and eventually fusion deficit of autophagosome and lysosome (Ding et al. Citation2020). Other studies reported that METH affected the expression of many lysosome proteins, such as lysosomal integral membrane protein type-2 (LIMP-2) (Li et al. Citation2018), microtubule-associated protein 1 light chain 3 (LC3) (Zhao et al. Citation2018), lysosome-associated membrane proteins (LAMPS), and suppressed the activity of cathepsin L (Nara et al. Citation2012). Combining our results with the literature findings, we believe that miRNAs could mediate the action of METH on lysosome. Furthermore, there are several of them related to nervous disorders. Rbm8a can promote neural stem cell proliferation and inhibit differentiation (Zou et al. Citation2015). SYT7 plays a key role in long-term plasticity (LTP) which is the basis of learning and memory (Glavan Citation2008). Overexpression of HTR1B could cause temporal lobe epilepsy (An and Kim Citation2011). RTN4 overexpression causes abnormalities in nerve signalling in schizophrenia (Berry et al. Citation2018). Overexpression of SV2A may lead to synaptic aberrations in hippocampal neurons (Nensa et al. Citation2014). Further study is needed to elucidate the role of miRNAs in regulating these putative target genes in response to METH exposure.
Conclusions
The miRNA profiles in the NAc of rats and the potential targets of the differential miRNAs indicated that the aberrance in miRNA expression was closely associated with METH exposure. These significantly changed miRNAs could be candidates for further functional studies on METH addiction. The bioinformatic analyses of the differential miRNAs suggested that these molecules may contribute to METH-induced alterations in the nervous system through the Wnt signalling pathway, axon guidance, and lysosome pathways. We deduced that the miRNAs mediated the action of METH on lysosomes in cells. Further studies on the function and regulatory mechanisms of these miRNAs are needed.
Supplementary_Table_S1.docx
Download MS Word (16 KB)Disclosure statement
The authors report no declarations of interest.
Additional information
Funding
References
- An SJ, Kim DS. 2011. Alterations in serotonin receptors and transporter immunoreactivities in the hippocampus in the rat unilateral hypoxic-induced epilepsy model. Cell Mol Neurobiol. 31:1245–1255.
- Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. 2006. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 124:191–205.
- Berry S, Weinmann O, Fritz AK, Rust R, Wolfer D, Schwab ME, Gerber U, Ster J. 2018. Loss of Nogo-A, encoded by the schizophrenia risk gene Rtn4, reduces mGlu3 expression and causes hyperexcitability in hippocampal CA3 circuits. PLoS One. 13:e0200896.
- Beveridge NJ, Tooney PA, Carroll AP, Gardiner E, Bowden N, Scott RJ, Tran N, Dedova I, Cairns MJ. 2008. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum Mol Genet. 17:1156–1168.
- Bosch PJ, Benton MC, Macartney-Coxson D, Kivell BM. 2015. mRNA and microRNA analysis reveals modulation of biochemical pathways related to addiction in the ventral tegmental area of methamphetamine self-administering rats. BMC Neurosci. 16:43.
- Brauer LH, de Wit H. 1996. Subjective responses to d-amphetamine alone and after pimozide pretreatment in normal, healthy volunteers. Biol Psychiatry. 39:26–32.
- Bu Q, Lv L, Yan G, Deng P, Wang Y, Zhou J, Yang Y, Li Y, Cen X. 2013. NMR-based metabonomic in hippocampus, nucleus accumbens and prefrontal cortex of methamphetamine-sensitized rats. Neurotoxicology. 36:17–23.
- Campos A, Pereira R, Vaz A, Caetano T, Malta M, Oliveira J, Carvalho FP, Mendo S, Lourenço J. 2020. Metals and low dose IR: molecular effects of combined exposures using HepG2 cells as a biological model. J Hazard Mater. 396:122634.
- Chandler RK, Finger MS, Farabee D, Schwartz RP, Condon T, Dunlap LJ, Zarkin GA, McCollister K, McDonald RD, Laska E, et al. 2016. The SOMATICS collaborative: introduction to a National Institute on Drug Abuse cooperative study of pharmacotherapy for opioid treatment in criminal justice settings. Contemp Clin Trials. 48:166–172.
- Chandrasekar V, Dreyer JL. 2009. microRNAs miR-124, let-7d and miR-181a regulate cocaine-induced plasticity. Mol Cell Neurosci. 42:350–362.
- Chandrasekar V, Dreyer JL. 2011. Regulation of MiR-124, Let-7d, and MiR-181a in the accumbens affects the expression, extinction, and reinstatement of cocaine-induced conditioned place preference. Neuropsychopharmacology. 36:1149–1164.
- Cook CE, Jeffcoat AR, Hill JM, Pugh DE, Patetta PK, Sadler BM, White WR, Perez-Reyes M. 1993. Pharmacokinetics of methamphetamine self-administered to human subjects by smoking S-(+)-methamphetamine hydrochloride. Drug Metab Dispos. 21:717–723.
- Di Chiara G. 2002. Nucleus accumbens shell and core dopamine: differential role in behavior and addiction. Behav Brain Res. 137:75–114.
- Ding J, Hu S, Meng Y, Li C, Huang J, He Y, Qiu P. 2020. Alpha-Synuclein deficiency ameliorates chronic methamphetamine induced neurodegeneration in mice. Toxicology. 438:152461.
- Fasciano J, Hatzidimitriou G, Yuan J, Katz JL, Ricaurte GA. 1997. N-methylation dissociates methamphetamine’s neurotoxic and behavioral pharmacologic effects. Brain Res. 771:115–120.
- Fiore R, Schratt G. 2007. MicroRNAs in synapse development: tiny molecules to remember. Expert Opin Biol Ther. 7:1823–1831.
- Glavan G. 2008. Intermittent L-DOPA treatment differentially alters synaptotagmin 4 and 7 gene expression in the striatum of hemiparkinsonian rats. Brain Res. 1236:216–224.
- Hamamoto DT, Rhodus NL. 2009. Methamphetamine abuse and dentistry. Oral Dis. 15:27–37.
- He Y, Yang C, Kirkmire CM, Wang ZJ. 2010. Regulation of opioid tolerance by let-7 family microRNA targeting the mu opioid receptor. J Neurosci. 30:10251–10258.
- Huang W, Li MD. 2009. Differential allelic expression of dopamine D1 receptor gene (DRD1) is modulated by microRNA miR-504. Biol Psychiatry. 65:702–705.
- Koob GF. 1992. Drugs of abuse: anatomy, pharmacology and function of reward pathways. Trends Pharmacol Sci. 13:177–184.
- Kosik KS. 2006. The neuronal microRNA system. Nat Rev Neurosci. 7:911–920.
- Larsen KE, Fon EA, Hastings TG, Edwards RH, Sulzer D. 2002. Methamphetamine-induced degeneration of dopaminergic neurons involves autophagy and upregulation of dopamine synthesis. J Neurosci. 22:8951–8960.
- Leshner AI, Koob GF. 1999. Drugs of abuse and the brain. Proc Assoc Am Physicians. 111:99–108.
- Li L, Chen S, Wang Y, Yue X, Xu J, Xie W, Qiu P, Liu C, Wang A, Wang H, et al. 2018. Role of GSK3β/α-synuclein axis in methamphetamine-induced neurotoxicity in PC12 cells. Toxicol Res (Camb). 7:221–234.
- Liang R, Lin Y, Ye JZ, Yan XX, Liu ZH, Li YQ, Luo XL, Ye HH. 2017. High expression of RBM8A predicts poor patient prognosis and promotes tumor progression in hepatocellular carcinoma. Oncol Rep. 37:2167–2176.
- Metzker ML. 2010. Sequencing technologies – the next generation. Nat Rev Genet. 11:31–46.
- Nakajima A, Yamada K, Nagai T, Uchiyama T, Miyamoto Y, Mamiya T, He J, Nitta A, Mizuno M, Tran MH, et al. 2004. Role of tumor necrosis factor-alpha in methamphetamine-induced drug dependence and neurotoxicity. J Neurosci. 24:2212–2225.
- Nara A, Aki T, Funakoshi T, Unuma K, Uemura K. 2012. Hyperstimulation of macropinocytosis leads to lysosomal dysfunction during exposure to methamphetamine in SH-SY5Y cells. Brain Res. 1466:1–14.
- Nensa FM, Neumann MHD, Schrötter A, Przyborski A, Mastalski T, Susdalzew S, Looβe C, Helling S, El Magraoui F, Erdmann R, et al. 2014. Amyloid beta a4 precursor protein-binding family B member 1 (FE65) interactomics revealed synaptic vesicle glycoprotein 2A (SV2A) and sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (SERCA2) as new binding proteins in the human brain. Mol Cell Proteomics. 13:475–488.
- Pietrzykowski AZ, Friesen RM, Martin GE, Puig SI, Nowak CL, Wynne PM, Siegelmann HT, Treistman SN. 2008. Posttranscriptional regulation of BK channel splice variant stability by miR-9 underlies neuroadaptation to alcohol. Neuron. 59:274–287.
- Reichel CM, Schwendt M, McGinty JF, Olive MF, See RE. 2011. Loss of object recognition memory produced by extended access to methamphetamine self-administration is reversed by positive allosteric modulation of metabotropic glutamate receptor 5. Neuropsychopharmacology. 36:782–792.
- Rodríguez-Feo JA, Puerto M, Fernández-Mena C, Verdejo C, Lara JM, Díaz-Sánchez M, Álvarez E, Vaquero J, Marín-Jiménez I, Bañares R, et al. 2015. A new role for reticulon-4B/NOGO-B in the intestinal epithelial barrier function and inflammatory bowel disease. Am J Physiol Gastrointest Liver Physiol. 308:G981–G993.
- Rogers JL, De Santis S, See RE. 2008. Extended methamphetamine self-administration enhances reinstatement of drug seeking and impairs novel object recognition in rats. Psychopharmacology (Berl). 199:615–624.
- Saba R, Störchel PH, Aksoy-Aksel A, Kepura F, Lippi G, Plant TD, Schratt GM. 2012. Dopamine-regulated microRNA MiR-181a controls GluA2 surface expression in hippocampal neurons. Mol Cell Biol. 32:619–632.
- Schratt G. 2009. microRNAs at the synapse. Nat Rev Neurosci. 10:842–849.
- Shi J, Zhou F, Wang LK, Wu GF. 2015. Synaptic vesicle protein2A decreases in amygdaloid-kindling pharmcoresistant epileptic rats. J Huazhong Univ Sci Technol [Med Sci]. 35:716–722.
- Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJL, Kane C, et al. 2009. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 11:705–716.
- Siomi H, Siomi MC. 2009. On the road to reading the RNA-interference code. Nature. 457:396–404.
- Wallace TL, Gudelsky GA, Vorhees CV. 1999. Methamphetamine-induced neurotoxicity alters locomotor activity, stereotypic behavior, and stimulated dopamine release in the rat. J Neurosci. 19:9141–9148.
- Wu Z, Sun Z, Huang R, Zang D, Wang C, Yan X, et al. 2020. Silencing of synaptotagmin 7 regulates osteosarcoma cell proliferation, apoptosis, and migration. Histol Histopathol. 35:303–312.
- Zajac MS, Renoir T, Perreau VM, Li S, Adams W, van den Buuse M, et al. 2018. Short-term environmental stimulation spatiotemporally modulates specific serotonin receptor gene expression and behavioral pharmacology in a sexually dimorphic manner in Huntington’s disease transgenic mice. Front Mol Neurosci. 11:433.
- Zhao C, Mei Y, Chen X, Jiang L, Jiang Y, Song X, Xiao H, Zhang J, Wang J. 2018. Autophagy plays a pro-survival role against methamphetamine-induced apoptosis in H9C2 cells. Toxicol Lett. 294:156–165.
- Zou D, McSweeney C, Sebastian A, Reynolds DJ, Dong F, Zhou Y, Deng D, Wang Y, Liu L, Zhu J, et al. 2015. A critical role of RBM8a in proliferation and differentiation of embryonic neural progenitors. Neural Dev. 10:18.