Abstract
Context
The potential hepatotoxicity of Polygoni Multiflori Radix (PMR) has attracted much attention, but the specific mechanism of inducing hepatotoxicity is still unclear due to the complexity of its components.
Objective
This study investigated the specific mechanism by which 2,3,5,4′-tetrahydroxy-stilbene-2-O-β-d-glucoside (TSG) regulates hepatotoxicity.
Materials and methods
The toxic effects of TSG (10, 100, 1000 μg/mL) on WRL-68 cells were examined using MTT, flow cytometry, and LDH assay after 24 h of incubation. Untreated cells served as the control. Gene and protein expression levels were determined by quantitative real-time PCR and Western blot, respectively. Immunofluorescence analysis was conducted to investigate the expression of light chain 3 (LC3). Luciferase activity assay was used to assess the targeted regulation of RUNX1 by miR-122.
Results
The half maximal inhibitory concentration (IC50) of TSG in WRL-68 cells was calculated as 1198.62 μg/mL. TSG (1000 μg/mL) inhibited cell viability and LDH activity and promoted WRL-68 cell apoptosis by inducing autophagy. Subsequent findings showed that TSG induced autophagy and promoted apoptosis in WRL-68 cells by downregulating the levels of p-PI3K, p-Akt, and p-mTOR proteins, while RUNX1 overexpression rescued this inhibition. Additionally, the effect of TSG on hepatocyte apoptosis was reversed by miR-122 knockdown. Furthermore, bioinformatics and dual luciferase reporter assay results indicated that miR-122 targeted RUNX1.
Discussion and conclusions
Our data demonstrate for the first time that TSG regulates hepatotoxicity, possibly by upregulating miR-122 and inhibiting the RUNX1-mediated PI3K/Akt/mTOR pathway to promote autophagy and induce hepatocyte apoptosis. Further in vivo research is necessary to verify our conclusion.
Keywords:
Introduction
Polygoni Multiflori Radix (PMR), which is derived from the roots of Polygonum multiflorum Thunb. (Polygonaceae), functions in detoxification, tonifying the liver and kidney, replenishing blood, etc. (Li, Wang, et al. Citation2017; Ruan et al. Citation2019). Although PMR has been widely applied in medical research, health care, and beauty products, reports on hepatotoxicity caused by PMR and related preparations have increased in recent years, and its safety has attracted wide attention (Lin et al. Citation2015). In addition, due to the complexity of its composition and function, the specific mechanism by which PMR causes hepatotoxicity is unclear.
The chemical composition of PMR is relatively complex, mainly containing stilbene, lecithin, anthraquinones, etc. Stilbene includes 2,3,5,4′-tetrahydroxy-stilbene-2-O-β-d-glucoside (TSG) (Ruan et al. Citation2019). TSG is the main active chemical constituent of PMR, which has therapeutic effects on the nervous, cardiovascular, digestive, endocrine, and respiratory systems (Xia et al. Citation2018). Recently, a great number of studies have shown that TSG may induce hepatocyte damage and cause liver damage (Xing et al. Citation2019). For example, TSG causes liver damage by inhibiting the mRNA expression of the cytochrome P450 enzyme CYP1A2, resulting in liver metabolic dysfunction (Hansen et al. Citation2019). In addition, studies also found that TSG does not cause abnormalities in related indicators of liver function, but it significantly damages the epithelial cells of the bile duct and interferes with liver cell function (Bommaya et al. Citation2011). Nevertheless, the specific mechanism by which TSG induces hepatotoxicity is unclear.
Autophagy, known as type II programmed cell death, degrades excess protein aggregates and dysfunctional organelles through lysosomes. Moderate autophagy maintains the stability of the internal environment, while excessive autophagy leads to cell death (Gross and Graef Citation2020). A key regulator of autophagy is the kinase target of rapamycin, which is a receptor for intracellular amino acids, ATP, and hormones. Rapamycin induces autophagy by inhibiting the mammalian TOR (mTOR) pathway (Zhang, Cai, et al. Citation2019). In recent years, the function of autophagy in drug-induced hepatotoxicity and other liver diseases has been reported. For example, FGF21 plays a protective role in acetaminophen-induced hepatotoxicity by activating autophagy in mice (Zhang et al. Citation2018). Glycine coumarin relieves alcohol-induced hepatotoxicity by inducing autophagy (Song et al. Citation2015). In addition, a study on the key pathway that mediates autophagy showed that the PI3K/Akt/mTOR pathway regulates aconitine-induced hepatocyte autophagy (Yang et al. Citation2019). At present, TSG-induced hepatotoxicity has been confirmed (Li, Wang, et al. Citation2017; Ruan et al. Citation2019). However, the specific mechanism of its induction of hepatotoxicity has not been reported. In particular, it is unclear whether TSG induces hepatocyte toxicity by modulating the PI3K/Akt/mTOR pathway.
MiRNA-122 (miR-122), which is a liver-specific conserved miRNA, regulates the metabolic homeostasis of hepatocytes and the replication process of hepatovirus and maintains balance in the liver (Chowdhary et al. Citation2017). Findings have shown that miR-122 is an early biomarker of liver cancer cells for the diagnosis and evaluation of early liver cancer (Amr et al. Citation2017). In addition, recent studies indicate that miR-122 may serve as a biomarker for drug-induced hepatocellular toxicity (Howell et al. Citation2017).
In this paper, we investigated the hepatotoxic effects of TSG with a focus on cell viability, cell apoptosis, and autophagy and dissected the potential molecular mechanisms responsible for the cytotoxic effects of TSG on hepatocytes. This study may provide a theoretical basis for the mechanism of TSG in hepatotoxicity.
Materials and methods
Cell culture and drug treatment
Hepatocyte WRL-68 cells were obtained from Cell Resource Center, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, P.R. China). Cells were cultured in DMEM-modified medium (Thermo Fisher Scientific, Waltham, MA, USA), which was supplemented with 10% FBS and 1% penicillin-streptomycin solution (Solarbio, Beijing, P.R. China), and cells were cultured at 37 °C in a humidified incubator containing 5% CO2. TSG was purchased from Nanjing Jingzhu Bio-technology Co., Ltd., (Nanjing, China). After treatment of WRL-68 cells with 10, 100, or 1000 μg/mL TSG, the effects of TSG on WRL-68 cytotoxicity, autophagy, and apoptosis were analysed by MTT assay, LDH assay, flow cytometry, etc. 3-MA and the PI3K inhibitor LY294002 were purchased from Sigma (St. Louis, MO, USA; assay >98%).
Plasmid vector construction and cell transfection
MiR-122 mimics, inhibitor and their respective negative controls (NC) with random sequences (denoted as mimics NC and inhibitor NC) were synthesized by RiboBio (Guangzhou, P.R. China). The full-length RUNX1 sequence was amplified and ligated into the pcDNA3.1 plasmid (OriGene Technologies, Inc.), and the recombinant plasmid was referred to as RUNX1. All plasmids were transfected by using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions in cells cultured in 6-well plates before the experiment. Cultured cells were collected for analysis following 48 h of transfection.
MTT assay
The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, Sigma, St. Louis, MO, USA) assay was performed to evaluate cell viability. In detail, 1 × 105 WRL-68 cells were seeded in 96‐well plates and incubated with 20 μL MTT reagent (5 mg/mL) for 4 h at 37 °C. After removing the medium, 150 μL of dimethyl sulfoxide (DMSO, Sigma, St. Louis, MO, USA) was added. A microplate reader (Bio‐Rad Laboratories, Hercules, CA, USA) was used to evaluate cell viability at 570 nm absorbance.
Cytotoxicity assay
After treatment with a certain concentration of TSG, the WRL-68 cell culture medium supernatant was harvested. According to the operating instructions, LDH levels were measured using an LDH cytotoxicity detection kit (Roche, Basel, Switzerland). The absorbance of the samples was measured using a microplate reader at a wavelength of 490 nm. Each sample was run in triplicate.
Flow cytometry analysis
The Annexin V-FITC/PI Apoptosis Detection Kit (Beyotime, Nanjing, P.R. China) was used for apoptosis analysis according to the manufacturer’s instructions. In detail, the 12-well cell culture plate was centrifuged at 1000 g for 5 min after the induction of apoptosis. The medium was aspirated, and the cells were washed twice with 1 × PBS for 5 min each time. Then, the cells were resuspended in 500 μL of binding buffer, and 5 μL of FITC-conjugated antibody and 5 μL of propidium iodide (PI) were added, followed by incubation for 15 min on ice in the dark. Finally, apoptosis was evaluated by flow cytometry (BD FACSCalibur; BD Biosciences) based on cell binding to Annexin V.
Immunofluorescence
Immunofluorescence was performed as described with a minor modification (Cui et al. Citation2019). In detail, 1 × 105 WRL-68 cells were seeded in 12-well plates for transfection. Cells were washed with 1 × PBS and fixed for 15 min in 4% formaldehyde. Then, Triton-X 100 was added to increase membrane permeability. After washing and blocking, WRL-68 cells were incubated with anti-LC3 antibody (Cell Signalling Technology, Danvers, MA, USA) overnight at 4 °C. After washing again, the cells were incubated with a secondary antibody for 1 h. Then, the cells were counterstained with DAPI, sealed with cover glass, and finally observed with a confocal fluorescence microscope (LSM880, Zeiss, Germany).
RNA extraction and qRT-PCR
TRIzol reagent (Invitrogen, Waltham, MA, USA) was used to extract total RNA from WRL-68 cells, and total RNA was reverse transcribed into complementary DNA (cDNA). Then qRT-PCR was performed by using SYBR Green PCR Master Mix (Takara, Dalian, P.R. China). For mRNA expression levels, β-actin served as a control; for miRNA expression, U6 served as a control. The 2−ΔΔCt method was used to calculate the relative expression. This experiment was repeated three more times. The primers used in this assay are displayed in .
Table 1. Primer sequences for quantitative real-time PCR.
Western blot
Western blotting was performed as previously described (Dong et al. Citation2017). Proteins were extracted from WRL-68 cells based on the provided instructions of the protein extraction kit (Beyotime, Nanjing, China). The protein samples were subjected to 10% SDS-PAGE and transferred to a PVDF membrane (PVDF, Millipore, Billerica, MA, USA). Then, the PVDF membrane was placed in 4% milk and incubated with the following primary antibodies (Abcam, Cambridge, UK): anti-LC3B (ab51520; 1:3000, detects LC3I and LC3II), anti-Beclin-1 (ab217179; 1:1000), anti-p62 (ab56416; 1:1000), anti-Bax (ab32503; 1:1000), anti-Bcl-2 (ab59348; 1:500), anti-cleaved caspase-3 (ab2302; 1:1000), anti-cleaved caspase-9 (ab2324; 1:1000) and anti-β-actin (ab8227; 1:5000) at 4 °C overnight. On the next day, the PVDF membrane was washed and incubated with a secondary antibody (Abcam, Cambridge, UK, 1:1000) for 1 h at room temperature. Finally, Pierce® ECL (Pierce, Rockford, IL, USA) Substrate A and B solutions were mixed in a 1:1 ratio and evenly distributed onto the PVDF membrane. The signals were visualized by Tanon 5200. Total protein levels were determined via ImageJ software (Media Cybernetics, Rockville, MD, USA) with β-actin as a normalization control.
Luciferase activity assay
The dual-luciferase reporter assay was conducted as previously described (Fu et al. Citation2019), with a slight modification. The wild-type or mutant (mut) RUNX1 sequence was amplified and cloned into the psi-CHECK2 reporter vector (Promega, Madison, WI, USA). A total of 2 × 104 293 T cells (ATCC, Manassas, VA, USA) were plated into 24-well plates and were subsequently co-transfected with the wild-type or mut RUNX1 3′-UTR vector and miR-122 mimics or NC mimics by using Lipofectamine® 2000 (Invitrogen, Waltham, MA, USA) at 37 °C for 48 h. According to the manufacturer’s protocol, the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) was used to detect the firefly luciferase activity of 293 T cells.
Statistical analysis
Data are presented as the mean ± SD of three independent experiments. Statistical analysis was performed using GraphPad Prism version 5.0 software (GraphPad Software, San Diego, CA). Differences between groups were analysed with Student’s t-test or one-way ANOVA followed by Tukey’s post hoc test for multiple comparisons. Differences were considered to be statistically significant at a p-value <0.05.
Results
TSG induces apoptosis and promotes autophagy in WRL-68 cells
TSG is an active ingredient extracted from the Chinese herbal medicine Polygonum multiflorum (Xia et al. Citation2018). Studies have shown that TSG induces hepatotoxicity and causes liver damage (Li, Niu, et al. Citation2017). To investigate the effect of TSG on hepatocyte WRL-68 cells, the cells were treated with 10, 100, or 1000 μg/mL TSG for 48 h, and the effects of TSG on biofunctions of WRL-68 cells were evaluated by MTT assay, LDH assay, etc. The MTT assay indicated that 10 and 100 μg/mL TSG had no effect on WRL-68 cell viability. However, 1000 μg/mL TSG inhibited the viability of WRL-68 cells compared with the control (). Similarly, TSG at 10 and 100 μg/mL had no effect on the LDH activity of WRL-68 cells, but TSG at 1000 μg/mL increased the LDH activity of WRL-68 cells (). As shown in , 1000 μg/mL TSG significantly promoted WRL-68 cell apoptosis compared with the control. In addition, immunofluorescence and Western blot analysis showed that TSG suppressed anti-apoptotic protein Bcl-2 expression but promoted cleaved-caspase 3, Bax, and cleaved-caspase 9 protein expression (). Accordingly, the autophagy-associated protein LC3 was significantly upregulated by TSG (). Based on these results, we conclude that TSG induces apoptosis and promotes autophagy in WRL-68 cells.
Figure 1. TSG induced apoptosis and promoted autophagy in WRL-68 cells. WRL-68 cells were treated with 10, 100 or 1000 μg/mL TSG, and untreated cells served as a control. Then, an MTT assay was performed to determine the effect of TSG on cell viability (A); LDH kits were used to detect LDH activity in WRL-68 cells (B); and cell apoptosis was evaluated by flow cytometry (C). WRL-68 cells were treated with 1000 μg/mL TSG, and untreated cells served as a control. Then, apoptosis-related protein expression was detected by Western blot (D); immunofluorescence was performed to detect LC3 protein expression and distribution (E; scale bar, 50 μm); and autophagy-related protein expression was detected by Western blot (F). Comparisons were performed using paired t-test or one-way ANOVA. * p < 0.05, ** p < 0.01. Error bars represent SD. Data represent three independent experiments.
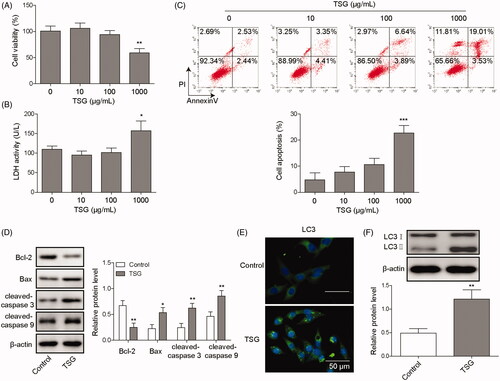
TSG promotes apoptosis by inducing autophagy in WRL-68 cells
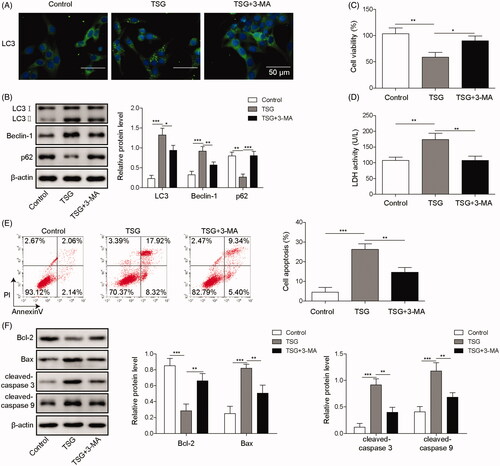
3-Methyladenine (3-MA) is an autophagy inhibitor that participates in the regulation of autophagy (Huang et al. Citation2019). To further investigate whether TSG promotes apoptosis by inducing autophagy in WRL-68 cells, WRL-68 cells were treated with 1000 μg/mL TSG and treated with 5 mM 3-MA for 1 h. Immunofluorescence and Western blot analysis indicated that the autophagy inhibitor 3-MA rescued WRL-68 cell autophagy induced by TSG (). As shown in , 3-MA increased WRL-68 cell viability inhibited by TSG. In addition, the LDH activity assay showed that 3-MA inhibited LDH activity in WRL-68 cells induced by TSG (). Similarly, 3-MA inhibited WRL-68 cell apoptosis induced by TSG (). Moreover, 3-MA promoted the expression of the anti-apoptotic protein Bcl-2 and inhibited Bax, cleaved-caspase 3, and cleaved-caspase 9 protein expression (). Based on these data, we suggest that TSG promotes apoptosis by inducing autophagy in WRL-68 cells.
Figure 2. TSG promoted apoptosis by inducing autophagy in WRL-68 cells. WRL-68 cells were treated with 1000 μg/mL TSG and 5 mM 3-MA, and untreated cells served as a control. Then, immunofluorescence was carried out to detect the effect of 3-MA on cell autophagy induced by TSG (A; scale bar, 50 μm); autophagy-related protein expression was measured by Western blot (B); MTT assay was carried out to determine the cell viability (C); LDH kits were used to detect LDH activity of WRL-68 cells (D); flow cytometry was performed to detect cell apoptosis (E); and Western blotting was performed to measure apoptosis-related protein expression (F). Comparisons were performed using paired t-test or one-way ANOVA. *p < 0.05, **p < 0.01, *** p < 0.001. Error bars represent SD. Data represent three independent experiments.
TSG induces autophagy and promotes apoptosis in WRL-68 cells by regulating the PI3K/Akt/mTOR pathway
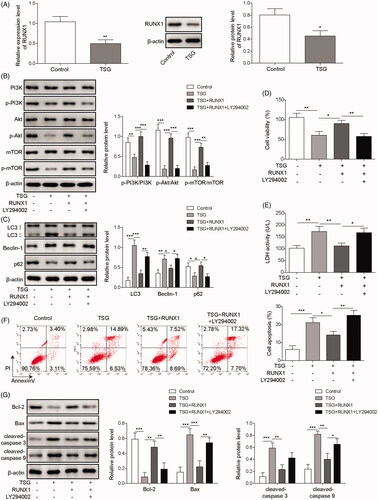
RUNX1 is involved in the regulation of the cell cycle and plays a vital role in tumour progression (Liu et al. Citation2020). To investigate whether RUNX1 regulated the apoptosis and autophagy of WRL-68 cells induced by TSG, Western blotting and qRT-PCR were performed to detect RUNX1 expression following 1000 μg/mL TSG treatment. The results indicated that RUNX1 expression was downregulated following treatment with TSG compared with the control (). Analysis of key proteins in the PI3K/Akt/mTOR pathway showed that TSG downregulated the expression of p-PI3K, p-Akt, and p-mTOR proteins, while overexpression of RUNX1 rescued this inhibition. In addition, LY294002, a PI3K inhibitor, reversed the effect of overexpressed RUNX1 (). As shown in , RUNX1 overexpression rescued WRL-68 cell autophagy induced by TSG, while LY294002 restored the autophagy effect. In addition, MTT assay indicated that overexpression of RUNX1 increased the viability of WRL-68 cells inhibited by TSG, while LY294002 attenuated this effect (). Similarly, overexpression of RUNX1 inhibited LDH activity in WRL-68 cells induced by TSG, while the effect was attenuated by LY294002 (). In terms of apoptosis, overexpression of RUNX1 inhibited TSG-induced apoptosis of WRL-68 cells, while LY294002 attenuated the inhibitory effect (). Consistently, overexpression of RUNX1 rescued the inhibitory effect of TSG on the anti-apoptosis protein Bcl-2 and attenuated the pro-apoptotic effect of TSG on the apoptosis-related proteins Bax, cleaved-caspase 3, and cleaved-caspase 9, while the above effect was attenuated by LY294002 (). Based on these results, we propose that TSG induces autophagy and promotes apoptosis in WRL-68 cells by regulating the PI3K/Akt/mTOR pathway.
Figure 3. TSG induced autophagy and promoted apoptosis in WRL-68 cells by regulating the PI3K/Akt/mTOR pathway. WRL-68 cells were treated with 1000 μg/mL TSG, and untreated cells served as a control. Then, mRNA and protein expression of RUNX1 was detected by qRT-PCR and Western blot (A). WRL-68 cells were transfected with the RUNX1-overexpressing plasmid for 48 h and then treated with 1000 μg/mL TSG following LY294002. Then, p-PI3K, p-Akt, and p-mTOR protein expression was detected by Western blot (B); Western blotting was carried out to measure autophagy-related protein expression (C); MTT assay was performed to determine cell viability (D); LDH kits were used to detect LDH activity of WRL-68 cells (E); cell apoptosis was evaluated by flow cytometry (F); and Western blotting was performed to detect apoptosis-related protein expression (G). Comparisons were performed using paired t-test or one-way ANOVA. *p < 0.05, **p < 0.01, ***p < 0.001. Error bars represent SD. Data represent three independent experiments.
TSG induces autophagy and promotes apoptosis in WRL-68 cells by upregulating miR-122 expression
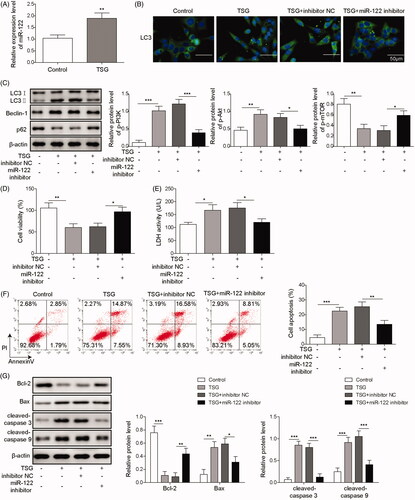
MiR-122 participates in the regulation of hepatocyte autophagy and apoptosis (Wei et al. Citation2019). To investigate the interaction between TSG, miR-122 and WRL-68 cell biological functions, miR-122 was inhibited in WRL-68 cells treated with TSG, and the autophagy and apoptosis of WRL-68 cells were detected by qRT-PCR, Western blot, etc. First, the effect of miR-122 on cell viability was evaluated by MTT assay. The results showed that knockdown of miR-122 inhibited WRL-68 cell viability, indicating that miR-122 inhibitor have a toxic effect on cells (Figure S1). Then qRT-PCR analysis results showed that TSG inhibited miR-122 expression (). Inhibition of miR-122 rescued WRL-68 cell autophagy induced by TSG but restored the dysregulation of autophagy-associated proteins induced by TSG (). In addition, the MTT assay showed that miR-122 inhibition increased WRL-68 cell viability inhibited by TSG (). As shown in , inhibition of miR-122 suppressed the LHD activity of WRL-68 cells induced by TSG. Similarly, inhibition of miR-122 inhibited the apoptosis of WRL-68 cells induced by TSG (). Similarly, inhibition of miR-122 restored the dysregulation of apoptosis-associated proteins induced by TSG. Based on these results, we conclude that TSG induces autophagy and promotes apoptosis in WRL-68 cells by upregulating miR-122 expression.
Figure 4. TSG induced autophagy and promoted apoptosis in WRL-68 cells by upregulating miR-122 expression. The miR-122 inhibitor was transfected into WRL-68 cells before treatment with 1000 μg/mL TSG. Then, qRT-PCR was performed to detect miR-122 expression (A); immunofluorescence was carried out to detect the influence of miR-122 on cell autophagy induced by TSG (B; scale bar, 50 μm); Western blotting was carried out to measure autophagy-related protein expression (C); MTT assay was performed to determine cell viability (D); LDH kits were used to detect LDH activity of WRL-68 cells (E); cell apoptosis was evaluated by flow cytometry (F); and Western blot was used to measure apoptosis-related protein expression (G). Comparisons were performed using paired t-test or one-way ANOVA. *p < 0.05, **p < 0.01, ***p < 0.001. Error bars represent SD. Data represent three independent experiments.
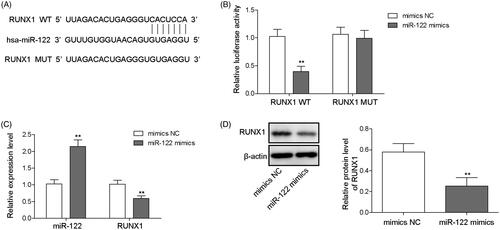
MiR-122 targets the regulation of RUNX1
MiR-122 participates in tumour progression in the liver by regulating target proteins (Amr et al. Citation2017; Cao and Yin Citation2019). To investigate the interaction between miR-122 and RUNX1, StarBase software was used to predict the potential target genes of miR-122. The findings indicated that the 3′-UTR of RUNX1 harboured a binding site of miR-122 (). To verify the binding of miR-122 and RUNX1, a dual luciferase reporter assay was performed. The data suggested that luciferase activity was clearly decreased after co-transfection of miR-122 mimics and the wild-type RUNX1 vector but was not altered after co-transfection of miR-122 mimics and the mutant RUNX1 vector (). In addition, qRT-PCR and Western blot analyses indicated that miR-122 overexpression inhibited RUNX1 expression in WRL-68 cells (). Therefore, we confirmed that miR-122 directly binds to RUNX1 and subsequently regulates its expression.
Figure 5. miR-122 targeted RUNX1. The potential binding site between miR-122 and RUNX1 was predicted by StarBase software (A); the direct binding relationship between miR-122 and RUNX1 was verified by the dual luciferase reporter assay (B); and the miR-122 mimics and luciferase reporter plasmids with wild-type or mutant RUNX1 3′-UTR were co-transfected into cells. Cells were transfected with NC mimics, miR-122 mimics, NC inhibitor, or miR-122 inhibitor, and then qRT-PCR was performed to evaluate miR-122 and RUNX1 expression (C). Western blotting was performed to analyse RUNX1 expression (D). Comparisons were performed using paired t-test or one-way ANOVA. *p < 0.05, **p < 0.01. Error bars represent SD. Data represent three independent experiments.
Discussion
Obtained from the roots of Polygonum multiflorum, PMR nourishes blood, kidneys, and liver and prevents hair from turning grey (Ma et al. Citation2015). TSG is a highly enriched styrene glycoside in PMR that has lipid and antioxidant activity. Studies have shown that TSG causes numerous toxic effects, such as nephrotoxicity, genotoxicity and hepatotoxicity (Li et al. Citation2013). In this paper, our findings indicate that TSG induces hepatocyte apoptosis and promotes autophagy.
Autophagy, as a regulatory mechanism of cell adaptation to survival, plays an important role in cellular metabolism (Gross and Graef Citation2020). To date, the role of autophagy in drug-induced hepatotoxicity and in other liver diseases has been reported. For example, in liver damage caused by ischemia-reperfusion, autophagosomes formed in liver cells are sites for eliminating ineffective intracellular components and protect liver cell damage caused by ischemia-reperfusion (Zhang, Liu, et al. Citation2019). On the other hand, concanavalin A activates autophagy and ruptures the lysosomal membrane by immunologically related GTPase, resulting in acute hepatitis (Tan et al. Citation2008). Our findings show that TSG promotes hepatocyte apoptosis by inducing autophagy, resulting in hepatotoxicity.
The PI3K/Akt/mTOR pathway is one of the vital pathways for autophagy regulation, and its regulatory function has been reported in chondrocytes, renal tubular epithelial cells and bladder cancer cells (Yan et al. Citation2019). Studies have shown that a great number of drugs induce autophagy by downregulating the PI3K/Akt/mTOR pathway. For example, with the increase in the concentration of acutine, p-PI3K, p-Akt and p-mTOR levels are significantly reduced, and autophagy is enhanced (Yang et al. Citation2019). In this study, our findings indicate that TSG induces autophagy and promotes hepatocyte apoptosis by inhibiting the PI3K/Akt/mTOR pathway.
As a liver-specific miRNA, miR-122 plays a vital role in regulating hepatocyte growth and proliferation, differentiation and regulation (Amr et al. Citation2017). Recently, findings have shown that miR-122 is related to various liver diseases, such as alcoholic liver disease, drug-induced liver damage (DILI), and chronic hepatitis C (Lu et al. Citation2019). In addition, studies indicate that the miR-122 expression level in serum serves as a prognostic biomarker for DILI (Brandenburger et al. Citation2018). In this paper, our findings indicate that under induction with TSG, miR-122 expression is upregulated in hepatocytes. In addition, upregulated miR-122 induces autophagy and promotes hepatocyte apoptosis. Thus, we suggest that TSG may promote autophagy and induce hepatocyte apoptosis by upregulating miR-122. Moreover, our data show that TSG inhibits the PI3K/Akt/mTOR pathway by downregulating RUNX1 expression, thereby inducing autophagy and promoting hepatocyte apoptosis, and RUNX1 is negatively regulated by miR-122. Thus, we propose that TSG regulates hepatotoxicity, possibly by upregulating miR-122 and inhibiting the RUNX1-mediated PI3K/Akt/mTOR pathway to promote autophagy and induce hepatocyte apoptosis.
Conclusions
The results indicate that regulation of the RUNX1-mediated PI3K/Akt/mTOR pathway might play a vital role in TSG-induced apoptosis of WRL-68 cells, which has implications for the hepatotoxicity of TSG. More in vivo studies are needed to confirm the findings of this study.
Author contributions
Guarantor of integrity of the entire study: Bing Dai; study concepts: Lei Yang; study design: Lei Yang; definition of intellectual content: Wei Xing; literature research: Meng-Jiao Liu; clinical studies: Wei Xing; experimental studies: Lin Tang; data acquisition: Meng-Jiao Liu; data analysis: Lu Wang; statistical analysis: Wang-Zhong Xiao; manuscript preparation: Meng-Jiao Liu; manuscript editing: Lu Wang; manuscript review: Bing Dai.
Supplementary_Figure_S1.tif
Download TIFF Image (1.1 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Amr KS, Elmawgoud Atia HA, Elazeem Elbnhawy RA, Ezzat WM. 2017. Early diagnostic evaluation of miR-122 and miR-224 as biomarkers for hepatocellular carcinoma. Genes Dis. 4:215–221.
- Bommaya G, Meran S, Krupa A, Phillips AO, Steadman R. 2011. Tumour necrosis factor-stimulated gene (TSG)-6 controls epithelial-mesenchymal transition of proximal tubular epithelial cells. Int J Biochem Cell Biol. 43:1739–1746.
- Brandenburger T, Salgado Somoza A, Devaux Y, Lorenzen JM. 2018. Noncoding RNAs in acute kidney injury. Kidney Int. 94:870–881.
- Cao F, Yin LX. 2019. miR-122 enhances sensitivity of hepatocellular carcinoma to oxaliplatin via inhibiting mdr1 by targeting Wnt/β-catenin pathway. Exp Mol Pathol. 106:34–43.
- Chowdhary V, Teng KY, Thakral S, Zhang B, Lin CH, Wani N, Bruschweiler-Li L, Zhang X, James L, Yang D, et al. 2017. miRNA-122 protects mice and human hepatocytes from acetaminophen toxicity by regulating cytochrome P450 family 1 subfamily a member 2 and family 2 subfamily e member 1 expression. Am J Pathol. 187:2758–2774.
- Cui JH, Dong SM, Chen CX, Xiao W, Cai QC, Zhang LD, He HJ, Zhang W, Zhang XW, Liu T, et al. 2019. Microplitis bicoloratus bracovirus modulates innate immune suppression through the EIF4e-EIF-4a axis in the insect Spodoptera litura. Dev Comp Immunol. 95:101–107.
- Dong SM, Cui JH, Zhang W, Zhang XW, Kou TC, Cai QC, Xu S, You S, Yu DS, Ding L, et al. 2017. Inhibition of translation initiation factor EIF4a is required for apoptosis mediated by Microplitis bicoloratus bracovirus. Arch Insect Biochem Physiol. 96:28–32.
- Fu Y, Wang C, Zhang D, Chu X, Zhang Y, Li J. 2019. miR-15b-5p ameliorated high glucose-induced podocyte injury through repressing apoptosis, oxidative stress, and inflammatory responses by targeting Sema3A. J Cell Physiol. 234:20869–20878.
- Gross A, Graef M. 2020. Mechanisms of autophagy in metabolic stress response. J Mol Biol. 432:28–52.
- Hansen J, Palmfeldt J, Pedersen KW, Funder AD, Frost L, Hasselstrøm JB, Jornil JR. 2019. Postmortem protein stability investigations of the human hepatic drug-metabolizing cytochrome P450 enzymes CYP1a2 and CYP3a4 using mass spectrometry. J Proteomics. 194:125–131.
- Howell LS, Ireland L, Park BK, Goldring CE. 2017. miR-122 and other micrornas as potential circulating biomarkers of drug-induced liver injury. Expert Rev Mol Diagnostics. 415:1–8.
- Huang YP, Huang YW, Hsiao YJ, Li SC, Hsu YH, Tsai CH. 2019. Autophagy is involved in assisting the replication of bamboo mosaic virus in Nicotiana benthamiana. J Exp Bot. 70:4657–4670.
- Li C, Niu M, Bai Z, Zhang C, Zhao Y, Li R, Tu C, Li H, Jing J, Meng Y, et al. 2017. Screening for main components associated with the idiosyncratic hepatotoxicity of a tonic herb, Polygonum multiflorum. Front Med. 11:253–265.
- Li H, Wang X, Liu Y, Pan D, Wang Y, Yang N, Xiang L, Cai X, Feng Y. 2017. Hepatoprotection and hepatotoxicity of heshouwu, a Chinese medicinal herb: context of the paradoxical effect. Food Chem Toxicol. 108:407–418.
- Li Q, Zuo W, Li F. 2013. Chelating ligand-mediated hydrothermal synthesis of samarium orthovanadate with decavanadate as vanadium source. ScientificWorldJournal. 2013:127816.
- Lin L, Ni B, Lin H, Zhang M, Li X, Yin X, Qu C, Ni J. 2015. Traditional usages, botany, phytochemistry, pharmacology and toxicology of Polygonum multiflorum Thunb.: a review. J Ethnopharmacol. 159:158–183.
- Liu S, Zhang Y, Huang C, Lin S. 2020. miR-215-5p is an anticancer gene in multiple myeloma by targeting runx1 and deactivating the PI3K/AKT/MTOR pathway. J Cell Biochem. 121:1475–1316.
- Lu X, Liu Y, Xuan W, Ye J, Yao H, Huang C, Li J. 2019. Circ_1639 induces cells inflammation responses by sponging miR-122 and regulating tnfrsf13c expression in alcoholic liver disease. Toxicol Lett. 314:89–97.
- Ma J, Zheng L, He YS, Li HJ. 2015. Hepatotoxic assessment of Polygoni multiflori radix extract and toxicokinetic study of stilbene glucoside and anthraquinones in rats. J Ethnopharmacol. 162:61–68.
- Ruan LY, Li MH, Xing YX, Hong W, Chen C, Chen JF, Xu H, Zhao WL, Wang JS. 2019. Hepatotoxicity and hepatoprotection of Polygonum multiflorum Thund. as two sides of the same biological coin. J Ethnopharmacol. 230:81–94.
- Song X, Yin S, Huo Y, Liang M, Fan L, Ye M, Hu H. 2015. Glycycoumarin ameliorates alcohol-induced hepatotoxicity via activation of nrf2 and autophagy. Free Radic Biol Med. 89:135–146.
- Tan TL, Fang N, Neo TL, Singh P, Zhang J, Zhou R, Koh CG, Chan V, Lim SG, Chen WN. 2008. Tac1 GTPase is activated by hepatitis B virus replication-involvement of HBX. Biochim Biophys Acta. 1783:360–374.
- Wei X, Liu H, Li X, Liu X. 2019. Over-expression of miR-122 promotes apoptosis of hepatocellular carcinoma via targeting tlr4. Ann Hepatol. 522:187–194.
- Xia W, Rui W, Zhao W, Sheng S, Lei L, Feng Y, Zhao S. 2018. Stable isotope labeling and 2,3,5,4'-tetrahydroxystilbene-2-O-β-D-glucopyranoside biosynthetic pathway characterization in Fallopia multiflora. Planta Med. 247:613–623.
- Xing Y, Wang L, Wang C, Zhang Y, Zhang Y, Hu L, Gao X, Han L, Yang W. 2019. Pharmacokinetic studies unveiled the drug-drug interaction between trans-2,3,5,4'-tetrahydroxystilbene-2-O-β-D-glucopyranoside and emodin that may contribute to the idiosyncratic hepatotoxicity of Polygoni Multiflori Radix. J Pharm Biomed Anal. 164:672–680.
- Yan S, Jiang C, Li H, Li D, Dong W. 2019. Fam3a protects chondrocytes against interleukin-1β-induced apoptosis through regulating PI3K/Akt/mTOR pathway. Biochem Biophys Res Commun. 516:209–214.
- Yang H, Wang H, Liu Y, Yang L, Sun L, Tian Y, Zhao B, Lu H. 2019. The PI3K/Akt/mTOR signaling pathway plays a role in regulating aconitine-induced autophagy in mouse liver. Res Vet Sci. 124:317–320.
- Zhang LD, Cai QC, Cui JH, Zhang W, Dong SM, Xiao W, Li J, Kou TC, Zhang XW, He HJ, et al. 2019. A secreted-Cu/Zn superoxide dismutase from Microplitis bicoloratus reduces reactive oxygen species triggered by symbiotic bracovirus. Dev Comp Immunol. 92:129–139.
- Zhang Y, Liu D, Hu H, Zhang P, Xie R, Cui W. 2019. HIF-1α/BNIP3 signaling pathway-induced-autophagy plays protective role during myocardial ischemia-reperfusion injury. Biomed Pharmacother. 120:109464.
- Zhang Y, Pan Y, Xiong R, Zheng J, Li Q, Zhang S, Li X, Pan X, Yang S. 2018. FGF21 mediates the protective effect of fenofibrate against acetaminophen-induced hepatotoxicity via activating autophagy in mice. Biochem Biophys Res Commun. 503:474–481.
