Abstract
Objectives: Rheumatoid Arthritis (RA) is the autoimmune disease representing the circadian variations of symptoms such as morning stiffness of joints or increased production of cytokines around midnight. Clock genes have been reported to affect on the pathogenesis of RA, however, the detailed relation between clock genes and disease activities of RA has remained unclear.
Methods: In this study, 15 RA patients treated with biological disease modifying anti-rheumatic drugs (bDMARDs) were enrolled (TNF inhibitor, 5; IL-6 inhibitor, 5; CTLA4-IgG, 5). Blood samples were collected from RA patients before treatment and at the study end-point fulfilling DAS28-ESR < 3.2. Total RNA was extracted from leukocytes to examine the expressions of the clock genes. We then evaluated the correlation of the clock gene expression with disease activity and the diagnostic values of the clock genes.
Results: The expressions of the clock genes were significantly modulated by bDMARDs treatments. Disease activities were significantly correlated with the clock genes expressions, and disease remission/low disease activity could be distinguished from moderate/high disease activity due to the sensitivities, the specificities and the areas under the curves of that.
Conclusion: The expressions of the clock genes in leukocytes could be useful as novel biomarkers predicting disease activities and therapeutic efficacies for bDMARDs in RA treatments.
Introduction
Rheumatoid arthritis (RA) is a chronic arthritis characterized by synovial inflammation and hyperplasia with pannus formation, resulting in joint destruction [Citation1]. Recently, remarkable features of RA, such as morning stiffness of joints and increased production of cytokines during midnight, have been discussed in relation to circadian rhythms [Citation2].
In mammalian species, the circadian rhythm is controlled by the network of clock genes; Period (Per), Cryptochrome (Cry), Brain and muscle arnt-like protein 1 (Bmal1), Circadian locomotor output cycles kaput (Clock), D site of the albumin promoter binding protein (Dbp), Hepatic leukemia factor (Hlf), Thyrotroph embryonic factor (Tef), E4-binding protein 4 (E4bp4), Retinoic-acid-receptor-related orphan receptor a (RORa) and Rev-erba [Citation3]. The transcription factor BMAL1 and CLOCK heterodimerize and bind to E-box element of the promoter regions of Per, Cry, Dbp, Hlf, Tef, E4bp4, Rora and Rev-erba, leading to their transcriptions. Then, PER and CRY heterodimerize and inhibit the activity of BMAL1/CLOCK heterodimer, successively, the transcriptions of Per and Cry are suppressed. DBP, HLF and TEF are collectively called as the proline and acidic amino acid-rich basic leucine zipper (PAR-bZIP) family and activate transcription of Per, while E4BP4 inhibits transcription of Per by binding to D-box element. RORα binds to REV-ERB/ROR response element (RRE) to activate the transcription of Bmal1, Clock and E4bp4, while REV-ERBα binds to RRE to inhibit the transcriptions of those [Citation4].
As described above, clock genes maintain an elaborate relationship with each other, and operate an order indispensable to keep the homeostasis of human body. Indeed, disorders of circadian operation have been reported to initiate disease onset including cancer, sleep disorder, cardiovascular disease, psychosis and metabolic disease [Citation5–7]. In colorectal carcinoma, expressions of Per2 is associated with tumor size and survival ratio [Citation8]. Although, lacking a rhythmical expression [Citation9], RORα is required for differentiation of T helper 17 cell [Citation10] and negatively regulates the inflammatory response through proinflammatory cytokine Interleukin (IL)-6 and IL-8 in rat smooth muscle cells [Citation11]. E4bp4 was also required for development of natural killer (NK) cell, and the subcutaneous cancer in Smad3 knockout mice was suppressed by activated NK cells through E4BP4-mediated pathway [Citation12,Citation13]. Moreover, a mutation of serine 662 amino acid into glycine 662 in PER was reported to be responsible for familial advanced sleep phase disorder [Citation14].
In the clinical field of RA treatment, three types of biological disease modifying anti-rheumatic drugs (bDMARDs) are widely used; adalimumab (ADA) and certolizumab pegol (CZP) bind to tumor necrosis factor (TNF)-α to inhibit the activity of that [Citation15], tocilizumab (TCZ) binds to IL-6 receptor to inhibit the intracellular signaling through glycoprotein 130 to Janus kinase-Signal transducer and activator of transcription [Citation16], and abatacept (ABT) inhibits the cluster of differentiation (CD) 28/CD80 or CD86 signaling of T cell as a cytotoxic T-lymphocyte antigen (CTLA) 4 – immnoglobulin (Ig) G fusion protein [Citation17]. And recently, treatments with TNF inhibitor (TNFi), TCZ and ABT were reported to improve sleep qualities of RA patients suffered with sleep disorder [Citation18–20].
In this study, we examined the disease activities and the expressions of the clock genes in leukocytes of RA patients before/after treatment with bDMARDs, and proposed, here, the clock genes as a novel biomarker predicting disease activity of RA and therapeutic efficacies of bDMARDs.
Materials and methods
Patients and ethics
The study group included six healthy controls (HC) and 15 RA patients treated with bDMARDs at Kobe Kaisei Hospital, Japan. Among them, five patients were treated with TNFi (two patients with ADA and three patients with CZP), five patient were with TCZ and five patients were with ABT. All patients fulfilled the 2010 American college of rheumatology classification criteria [Citation21]. Blood samples were collected before treatments and at the study end-point achieving disease activity score in 28-joints erythrocyte sedimentation rate (DAS28-ESR)< 3.2. DAS28-ESR is determined by the formula [Citation22]; DAS28-ESR = 0.56*(square root of tender joints counts 28) + 0.28*(square root of swollen joints count 28) + 0.7*(natural logarithm of ESR) + .014*visual analog scale. The profile of HC was as follows; n = 6 (female ratio 66.7%) and age = 57.8 ± 6.0.
This study was approved by the ethics committee of Kobe University (approval #579) and Kobe Kaisei Hospital (approval #0017), and written informed consent was obtained from each patient before study enrolment, according to the Declaration of Helsinki.
Cell culuture, RNA extraction, reverse transcription, quantitative polymerase chain reaction
THP-1 cells were purchased from JCRB Cell Bank of National Institutes of Biomedical Innovation, Health and Nutrition (JCRB0112). Cells were maintained in RPMI1640 medium (Wako, Tokyo, Japan) supplemented with 10% of heat-inactivated fetal bovine serum, 1% penicillin–streptomycin (Life Technologies) at 37 °C and 5%CO2.
THP-1 cells were cultured (5.0 × 105 cells/mL) for 0–32 h, and stimulated with or without IL-6(50 ng/mL, Peprotech, Rocky Hill, NJ) or TNF-α(10 ng/mL, R&D systems, McKinley, NE) for additional 24 h. Total RNA was extracted by RNeasy Mini Kit and QIAshredder (Qiagen, Hilden, Germany), and by PAXgene Blood RNA kit (QIAGEN, Hilden, Germany) from blood samples. Reverse transcription was performed with ReverTraAce (TOYOBO, Osaka, Japan) to analyze the expressions of the clock genes in leukocyte by StepOnePlus Real-Time PCRSystems (Applied Biosystems, Foster City, CA). The TaqMan probes used were: Per2 (Hs00256143_m1), Cry1 (Hs00172734_m1), Cry2 (Hs00323654_m1), Clock (Hs00231857_m1), E4bp4 (Hs00993282_m1), Rora (Hs00536545_m1), and Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) (Hs99999905_m1).
Statistical analysis
Student’s t test was performed to compare the expressions of the clock genes between HC and patients’ samples (before-treatment or before and after-treatment), and that of THP-1 cells. For categorical variables, Fisher’s exact test was conducted. Kruskal–wallis test was used to evaluate the differences of clinical data among three types of bDMARDs. To examine the correlation between 2 independent values, Pearson’s correlation coefficients (shown as ‘r’ in the figures) and p-values were used. To evaluate the diagnostic values, area under receiver operating characteristic curve (AUC-ROC) was adapted. The cutoff values were determined by the points on the ROC with minimum distance from the upper-left corner of the graph.
All statistical tests were two-sided and p < .05 was considered as statistically significant. The statistical analysis were performed by EZR software version 1.36, based on R and R commander [Citation23].
Results
Profiles of patients with RA before/after bDMARDs treatment
The clinical characteristic and disease activity of 15 patients with RA were shown in . When comparing the patient background, methylprednisolone (PSL)-dosage, methotrexate (MTX)-dosage and other bDMARDs-use were significantly different among three groups; p = .033, p = .039 and p = .0093, respectively. Finally, DAS28-ESR in baseline and DAS28-ESR after treatment showed significant differences among three groups; p = .042 and p = .0030, respectively.
Table 1. Clinical informations of RA patients.
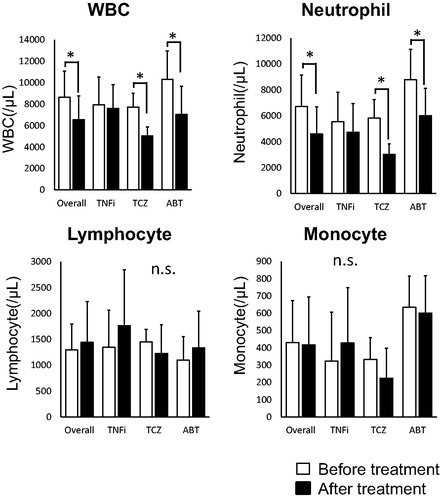
The leukocyte’ populations of RA patients were shown in . Leukocytes significantly decreased in TCZ-treated (p < .05), ABT-treated (p < .05) and overall patients (p < .01). Neutrophils significantly decreased in TCZ-treated (p < .05), ABT-treated (p < .05) and overall patients (p < .001).
The expressions of clock genes in leukocytes before/after bDMARDs treatment
We next studied the expressions of the circadian clock genes in leukocyte of HC and patients before/after treated with bDMARDs.
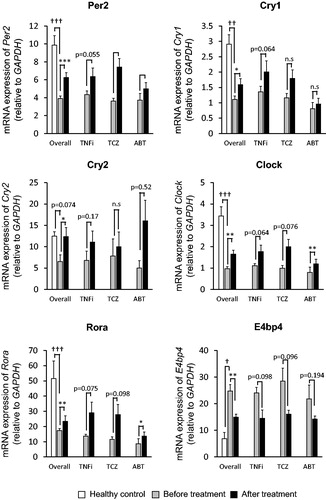
As shown in , the expressions of Per2, Cry1, Cry2, Clock and Rora in RA patients was lower than healthy controls; p < .000001, p < .00001, p = .0745, p < .000001, and p < .000001, respectively. While the expression of E4bp4 in RA patients was higher than healthy controls (p < .001). After treatments, expressions of Per2 were significantly increased in TCZ-treated (p < .05), ABT-treated (p < .01) and overall patients (p < .001), though was not significant in TNFi-treated patients (p = .055). Expression of Cry1 and Cry2 were significantly increased in overall patients (p < .05), though there was no significant difference for individual agents. Expressions of Clock were significantly increased in ABT-treated (p < .01) and overall patients (p < .01), and also they were increased in TNFi-treated and TCZ-treated patients though not statistically significant (p = .064 and .076, respectively). Expressions of Rora were increased in ABT-treated (p < .05) and overall patients (p < .01), and also increased in TNFi-treated and TCZ-treated patients (p = .075 and .098, respectively). Finally, Expressions of E4bp4 were decreased in overall patients (p < .01), though not significant in individual agents.
The correlations between DAS28-ESR and the clock genes in RA patients
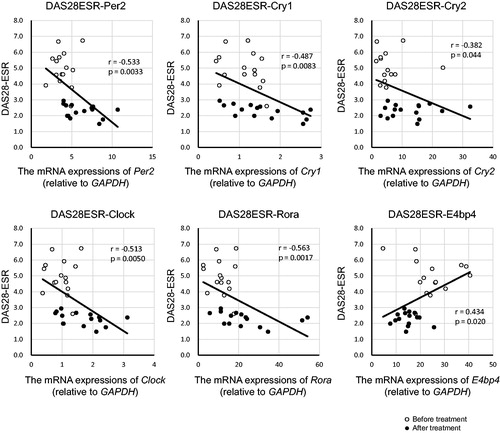
We next analyzed the correlation between DAS28-ESR and the expressions of the clock genes (). DAS28-ESR was negatively correlated with the expressions of Per2 (r = –.533, p < .01), Cry1 (r = –.487, p < .01), Cry2 (r = –.382, p < .05), Clock (r = –.513, p < .01) and Rora (r = –.563, p < .01), while was positively correlated with E4bp4 (r = 0.434, p < .05).
The cutoff values, the sensitivities, the specificities and AUCs of the clock genes in RA patients
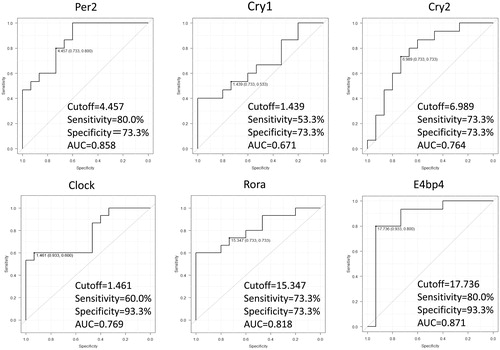
ROC curve analysis was performed to discriminate MDA/HDA from disease remission/LDA in RA patients by the expressions of the clock genes. As shown in , the diagnostic power of Per2 with a cutoff of 4.457 showed 80.0% sensitivity and 73.3% specificity, and AUC was 0.858. That of Cry1 with a cutoff of 1.439 showed 53.3% sensitivity, 73.3% specificity, and AUC was 0.671. That of Cry2 with a cutoff of 6.989 showed 73.3% sensitivity, 73.3% specificity, and AUC was 0.764. Also, that of Clock showed 60.0% sensitivity, 93.3% specificity, AUC 0.769 with a cutoff of 1.461. That of Rora with a cutoff of 15.35 showed 73.3% sensitivity and 73.3% specificity, and AUC was 0.818. At last, that of E4bp4 with a cutoff of 17.74 showed 80.0% sensitivity and 93.3% specificity, and AUC was 0.871.
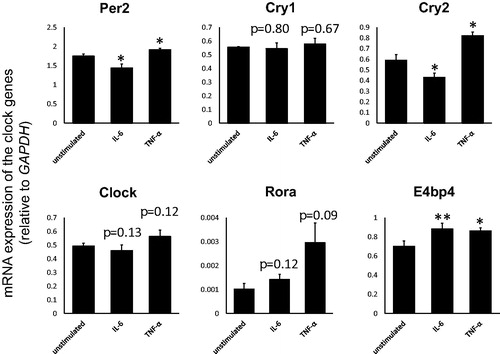
The expressions of the clock genes in THP-1
To explore the interaction of the clock genes in leukocytes, we next investigated the modulated expressions of the clock genes in THP-1 cells stimulated with IL-6 and TNF-α. As shown in , expressions of Per2 and Cry2 were significantly decreased by IL-6, while they were significantly increased by TNF-α. The expression of E4bp4 was significantly increased by both IL-6 and TNF-α.
Discussion
We previously reported that deletions of Cry1/2 gene exacerbated arthritis in mice due to excessive production of TNF-α from lymphocytes [Citation24] , on the other hand, stimulations of TNF-α modulated the expressions of the clock genes in RA fibroblast-like synoviocytes [Citation25,Citation26]. Indeed, treatments with another TNF-α inhibitory bDMARDs, infliximab, significantly modulated the expressions of Per1/2 and Cry2 in RA patients’ peripheral blood mononuclear cells [Citation27]. Journiac et al. have been also reported that RORα promotes IL-6 production by binding the promotor region of IL-6 [Citation28].Thus, the relationship between clock genes and inflammatory cytokines appears to be solidly connected and it is quite reasonable to conceive that the elevated cytokines’ concentration during midnight may be aggravating joint symptoms in the morning.
Since most of clock genes individually inherent their rhythm, expressions of those have been evaluated by discussing both expression level and expression phase. Per1 and Per2 genes rhythmically express in mouse peripheral leukocyte, as well as Per1/2 and Bmal1 in human peripheral leukocytes [Citation29]. So, it has been considered that a fixed-point observation does not necessarily reflect the characteristics of clock genes.
However, in this study, we collected patients’ blood samples at arbitrary timing during the outpatient clinic.
As shown in , the counts of WBC and neutrophil were decreased in TCZ-treated patients, consisting with a previous research [Citation30]. And given and Citation4, we could approximately speculate disease activity and distinguish disease remission/LDA from MDA/HDA in RA patients with high sensitivity and specificity with conveniently examining the expression levels of the clock genes in leukocytes.
When investigating the interaction of clock genes in leukocytes, as referred to and , expressions of Per2, Cry2, E4bp4 were consistent in both in vivo and in vitro, suggesting that expressions of Per2 and Cry2 could be solely modulated by IL-6 and E4bp4 could be modulated by both IL-6 and TNF-α in the monocytes. Further studies for the detailed relation among clock genes especially in cases with RA patients should be required.
At the present time, DAS28-ESR is a universally used index to estimate medical conditions of RA patients [Citation31]. Also, multi-biomarker disease activity (MBDA) was recently reported as a novel disease-assessing method correlated with DAS28 while it is a merged evaluation of 12 molecules in sera [Citation32]. In addition to these indications, nevertheless types of bDMARDs and timing of sample collection, we simply propose that expressions of the clock genes, especially Per2, E4bp4 and Rora, are also useful as biomarkers predicting the disease activity and the efficacies of bDMARDs in RA.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Conflict of interest
None.
Additional information
Funding
References
- Smolen JS, Aletaha D, McInnes IB. Rheumatoid arthritis. Lancet. 2016;388(10055):2023–38.
- Yoshida K, Hashimoto T, Sakai Y, Hashiramoto A. Involvement of the circadian rhythm and inflammatory cytokines in the pathogenesis of rheumatoid arthritis. J Immunol Res. 2014;2014:1.
- Gibbs JE, Ray DW. The role of the circadian clock in rheumatoid arthritis. Arthritis Res Ther. 2013;15(1):205.
- Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37(2):187–92.
- Maury E, Ramsey KM, Bass J. Circadian rhythms and metabolic syndrome: from experimental genetics to human disease. Circ Res. 2010;106(3):447–62.
- Lamont EW, Coutu DL, Cermakian N, Boivin DB. Circadian rhythms and clock genes in psychotic disorders. Isr J Psychiatry Relat Sci. 2010;47(1):27–35.
- Takeda N, Maemura K. Circadian clock and cardiovascular disease. J Cardiol. 2011;57(3):249–56.
- Momma T, Okayama H, Saitou M, Sugeno H, Yoshimoto N, Takebayashi Y, et al. Expression of circadian clock genes in human colorectal adenoma and carcinoma. Oncol Lett. 2017;14:5319–25.
- Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;12:929–37.
- Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28(1):29–39.
- Delerive P, Monté D, Dubois G, Trottein F, Fruchart-Najib J, Mariani J, et al. The orphan nuclear receptor ROR alpha is a negative regulator of the inflammatory response. EMBO Rep. 2001;2(1):42–8.
- Kamizono S, Duncan GS, Seidel MG, Morimoto A, Hamada K, Grosveld G, et al. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J Exp Med. 2009;206(13):2977–86.
- Tang PM, Zhou S, Meng XM, Wang QM, Li CJ, Lian GY, et al. Smad3 promotes cancer progression by inhibiting E4BP4-mediated NK cell development. Nat Commun. 2017;6:14677–91.
- Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, et al. An hPer2 hosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291(5506):1040–3.
- Radner H, Aletaha D. Anti-TNF in rheumatoid arthritis: an overview. Wien Med Wochenschr. 2015;165(1–2):3–9.
- Jones SA, Scheller J, Rose-John S. Therapeutic strategies for the clinical blockade of IL-6/gp130 signaling. J Clin Invest. 2011;121(9):3375–83.
- Blair HA, Deeks ED. Abatacept: a review in rheumatoid arthritis. Drugs. 2017;77(11):1221–33.
- Taylor-Gjevre RM, Gjevre JA, Nair BV, Skomro RP, Lim HJ. Improved sleep efficiency after anti-tumor necrosis factor α therapy in rheumatoid arthritis patients. Ther Adv Musculoskelet Dis. 2011;3(5):227–33.
- Fragiadaki K, Tektonidou MG, Konsta M, Chrousos GP, Sfikakis PP. Sleep disturbances and interleukin 6 receptor inhibition in rheumatoid arthritis. J Rheumatol. 2012;39(1):60–2.
- Wells G, Li T, Tugwell P. Investigation into the impact of abatacept on sleep quality in patients with rheumatoid arthritis, and the validity of the MOS-Sleep questionnaire Sleep Disturbance Scale. Ann Rheum Dis. 2010;69(10):1768–73.
- Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO III, et al. Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Atrhritis Rheum. 2010;l62:2569–81.
- Carpenter L, Norton S, Nikiphorou E, Kiely P, Walsh DA, Dixey J, et al. Validation of methods for converting the original Disease Activity Score (DAS) to the DAS28. Rheumatol Int. 2018;38(12):2297–305.
- Kanda Y. Investigation of the freely available easy-to-use software 'EZR' for medical statistics. Bone Marrow Transplant. 2013;48(3):452–8.
- Hashiramoto A, Yamane T, Tsumiyama K, Yoshida K, Komai K, Yamada H, et al. Mammalian clock gene Cryptochrome regulates arthritis via proinflammatory cytokine TNF-alpha. J Immunol. 2010;184(3):1560–5.
- Yoshida K, Hashiramoto A, Okano T, Yamane T, Shibanuma N, Shiozawa S. TNF-α modulates expression of the circadian clock gene Per2 in rheumatoid synovial cells. Scand J Rheumatol. 2013;42(4):276–80.
- Yoshida K, Nakai A, Kaneshiro K, Hashimoto N, Suzuki K, Uchida K, et al. TNF-α induces expression of the circadian clock gene Bmal1 via dual calcium-dependent pathways in rheumatoid synovial cells. Biochem Biophys Res Commun. 2018;495(2):1675–80.
- Yoshida K, Hashimoto T, Sakai Y, Hashiramoto A. Circadian rhythm joint stiffness destruction rheumatoid arthritis. Int J Clin Rheumtol. 2015;10(5):335–44.
- Journiac N, Jolly S, Jarvis C, Gautheron V, Rogard M, Trembleau A, et al. The nuclear receptor ROR(alpha) exerts a bi-directional regulation of IL-6 in resting and reactive astrocytes. Proc Natl Acad Sci USA. 2009;106(50):21365–70.
- Fukuya H, Emoto N, Nonaka H, Yagita K, Okamura H, Yokoyama M. Circadian expression of clock genes in human peripheral leukocytes. Biochem Biophys Res Commun. 2007;354(4):924–8.
- Wright HL, Cross AL, Edwards SW, Moots RJ. Effects of IL-6 and IL-6 blockade on neutrophil function in vitro and in vivo. Rheumatology (Oxford). 2014;53(7):1321–31.
- van der Heijde DM, van 't Hof MA, van Riel PL, Theunisse LA, Lubberts EW, van Leeuwen MA, et al. Judging disease activity in clinical practice in rheumatoid arthritis: first step in the development of a disease activity score. Ann Rheum Dis. 1990;49(11):916–20.
- Hirata S, Li W, Defranoux N, Cavet G, Bolce R, Yamaoka K, et al. A multi-biomarker disease activity score tracks clinical response consistently in patients with rheumatoid arthritis treated with different anti-tumor necrosis factor therapies: a retrospective observational study. Mod Rheumatol. 2015;25(3):344–9.