?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Astrocytes play an important role in the central nervous system, contributing to the development of and maintenance of synapses, recycling of neurotransmitters, and the integrity and function of the blood–brain barrier. Astrocytes are also linked to the pathophysiology of various neurodegenerative diseases. Astrocyte function and organization are tightly regulated by interactions mediated by the extracellular matrix (ECM). Engineered hydrogels can mimic key aspects of the ECM and can allow for systematic studies of ECM-related factors that govern astrocyte behaviour. In this study, we explore the interactions between neuroblastoma (SH-SY5Y) and glioblastoma (U87) cell lines and human fetal primary astrocytes (FPA) with a modular hyaluronan-based hydrogel system. Morphological analysis reveals that FPA have a higher degree of interactions with the hyaluronan-based gels compared to the cell lines. This interaction is enhanced by conjugation of cell-adhesion peptides (cRGD and IKVAV) to the hyaluronan backbone. These effects are retained and pronounced in 3D bioprinted structures. Bioprinted FPA using cRGD functionalized hyaluronan show extensive and defined protrusions and multiple connections between neighboring cells. Possibilities to tailor and optimize astrocyte-compatible ECM-mimicking hydrogels that can be processed by means of additive biofabrication can facilitate the development of advanced tissue and disease models of the central nervous system.
1. Introduction
Of all the cells that make up the human brain, astrocytes are the most abundant cell type. Astrocytes are involved in synapse formation and function, as well as metabolic activities to support neurons through glutamate clearance at the synaptic cleft [Citation1–3]. Astrocytes also constitute a key role in the formation of the gliovascular unit, where they interact with brain endothelial cells, pericytes, and vascular smooth muscle cells as well as extracellular matrix (ECM) components to form the blood–brain barrier (BBB). The BBB regulates the homeostasis of the central nervous system by controlling transport and exchange of molecules between the blood and the brain [Citation4]. In addition to cell–cell interactions, astrocytes interact extensively with the ECM and the gliovascular basal lamina components of the BBB [Citation5]. The basal lamina is rich in laminins and primarily interacts with the astrocytic endfeet [Citation6]. The interactions between astrocytes and the ECM are also thought to influence the astrocytic response to injury, inflammation, and disease [Citation7].
Astrocytes are highly involved in several pathological conditions of the central nervous system. For example, they act protectively by producing and secreting the antioxidant glutathione during oxidative stress, which is a common trait in neurodegenerative diseases [Citation8–10]. The size and shape of astrocytes also vary during different disease conditions. When astrocytes were studied in an epileptic model, they extended thicker and longer protrusions than healthy cells [Citation11]. During traumatic brain injury, where tissue lesions were formed, astrocytes were seen to extend long processes towards the injured area [Citation12]. Such lesions have also been studied in three-dimensional (3D) hydrogel models based on alginate, where changes in glial marker GFAP (glial fibrillary acidic protein) expression were observed when cultured in a meningeal fibroblast conditioned medium [Citation13]. The multifaceted interactions between astrocytes and the ECM must be carefully considered to generate biologically relevant hydrogel based 3D in vitro models of the BBB and other structures of the central nervous system, in addition to enable culture of neurons. To further facilitate the development of more sophisticated models with defined structures and spatial control over hydrogel composition and cell distribution, hydrogels that are compatible with advanced biofabrication techniques, such as extrusion bioprinting, should be used. In this work, we present a modular and bioprintable hyaluronan (HA) hydrogel system that allows for tuning of astrocyte–hydrogel interactions by providing flexible click-chemistry mediated possibilities to conjugate peptide cell adhesion motifs (cRGD and IKVAV; see below). The hydrogels, with and without cell adhesion motifs, were assessed using the 3D culture of the widely used human neuroblastoma model SH-SY5Y, the glioblastoma cell line U87, and human fetal primary astrocytes (FPA), and were further optimized to function as a bioink for 3D bioprinting of FPA.
The short peptide motifs Arg-Gly-Asp (RGD) and Ile-Lys-Val-Ala-Val (IKVAV) are derived from ECM proteins, such as fibronectin and laminins, and have been shown to improve cellular attachment in synthetic ECM models [Citation14]. Integration of linear or cyclic RGD (cRGD) has previously been explored by us using a HA-based gel to better mimic the in vivo environment for the 3D culture of a wide range of cell types, including hepatoma cells and human induced pluripotent stem cell-derived hepatocytes [Citation15]. Previous studies show that both RGD and IKVAV improved the ability of neural progenitor cells to differentiate into neurons and promotes neurite outgrowth [Citation16,Citation17]. It has also been demonstrated that IKVAV functionalized hydrogels promote neural differentiation over astrocyte differentiation of neural stem cells [Citation18]. The same study also shows how the IKVAV hydrogel successfully supports neural differentiation and tissue regeneration of the neural stem cells that were implanted in rat brains. HA-containing RGD-functionalized hydrogels have also been used to investigate how the microenvironment contributes to cell–ECM interactions and the survival of malignant brain tumor cells [Citation19]. By blocking the overexpressed HA receptor CD44 in the tumour cells, their adhesion to the ECM was reduced. Combined, these findings motivate further detailed investigations of the mechanism of interactions between neurons and astrocytes with ECM mimetic hydrogels. Hydrogel systems that both can provide a biologically relevant microenvironment and that are compatible with 3D bioprinting opens for possibilities to start replicating the heterogeneous composition and spatial complexity of structures in the brain, which is central for generating more advanced tissue and disease models.
2. Experimental section
2.1. Synthesis of HA-BCN
HA (150 kDa) obtained from Lifecore Biomedical (Minnesota, U.S.A) was modified with bicyclo[6.1.0]nonyne (BCN) as described in further detail in Selegård et al. [Citation20]. In brief, N-[(1 R,8 S,9 S)-Bicyclo[6.1.0]non-4-yn-9-ylmethyloxycarbonyl]-1,8diamino-3,6-dioxaoctane (BCN-NH2) was dissolved in 5:1 (v/v) acetonitrile:MilliQ water prior to adding 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride and 1-hydroxybenzotriazole hydrate. The mixture was then added to HA previously dissolved in 2-(N-morpholino)ethanesulfonic acid buffer (100 mM, pH 7). The carbodiimide reaction was allowed to continue for 24 h on a shaker at room temperature. Subsequently, time dialysis (molecular weight cutoff 12–14 kDa, Spectra/Por RC, Spectrum Laboratories Inc.) was performed in acetonitrile:MilliQ water (1:10 v/v) for 24 h and then for 3 days in MilliQ water. The final product, denoted as HA-BCN, was lyophilized and stored at −20°C. Based on the proton nuclear magnetic resonance (1 H-NMR) analysis, the HA-BCN had a derivatization degree of approximately 19%.
2.2. Peptide synthesis
The peptide cRGD with the sequence c(RGDfK(N3) was synthesized as described previously [Citation11]. The peptide IKVAV with the sequence K(N3)GGIKVAV-NH2 was synthesized on a Liberty Blue peptide synthesizer (CEM, Matthews, U.S.A) using standard fluorenylmethoxycarbonyl protecting group (Fmoc) chemistry. ProTide Rink Amide (0.19 mmol/g) was used as solid support, and the peptide was synthesized in a 100 μmol scale. Each coupling was performed under microwave conditions at 90°C for 2 min using a fivefold excess of amino acid and N,N-diisopropylcarbodiimid as coupling reagent and tenfold excess of Oxyma Pure as base. Fmoc deprotection was also performed under microwave conditions at 90°C for 1 min using 20% piperidine in dimethylformamide (DMF) (v/v). The peptide was, after final Fmoc deprotection, cleaved from its solid support by treatment with trifluoroacetic acid (TFA):H2O:trisioproylsilane (95:2.5:2.5, v/v/v) for 3 h before being concentrated and precipitated using ice-cold diethyl ether. The crude peptide was purified with a ReproSil gold C-18 column attached to a semi-preparative high-performance liquid chromatography system (HPLC; UltiMate 3000, Dionex, Sunnyvale, U.S.A) using an aqueous gradient of acetonitrile with 0.1% TFA. The identity of the peptide was confirmed using a matrix-assisted laser desorption time-of-flight mass spectrometer (MALDI-ToF-MS; UltrafleXtreme, Bruker, Billerica, U.S.A) running in positive ion mode with alpha-cyano-4-hydroxycinnamic acid as matrix (Figure S2). Peptide purity was confirmed using HPLC (Figure S3).
2.3. Hydrogel formation
HA-BCN and 8-arm azide-terminated polyglycol ethylene((PEG-Az)8) was suspended in cell media (including, where relevant, cells) unless stated otherwise to give a concentration of 1% (w/v). 10 mM peptide solution was added to the HA-BCN as 10% of the total hydrogel volume (i.e. 1 mM final concentration) and incubated for 1 hour at 37°C to allow for HA functionalization via strain-promoted alkyne-azide cycloadditions (SPAAC). The same volume of PBS was added instead of the peptide in the no peptide condition. HA-BCN + peptide and (PEG-Az)8 were combined at a ratio of 3:1, quickly but thoroughly mixed, and the gels formed as required. The plate was sealed, and incubation was performed for 1 h at 37°C to cross-link the gels, after which pre-warmed cell media was added to the wells.
2.4. Rheology
Oscillatory rheology was performed using a Discovery HR-2 rheometer (TA Instruments, U.S.A). Hydrogels were produced with a total volume of 30 µl as described above. Instead of cell media, phosphate-buffered saline (PBS) was added to completely cover the hydrogels and the incubation continued for further 24 h at 37°C. The hydrogels were analyzed using strain sweeps (1 Hz, 0.1% to 10% strain) and frequency sweeps (1% strain, 0.1 to 10 Hz) using an 8 mm parallel geometry. Gelation kinetics were performed using 50 µl volumes and a 20 mm 1° geometry at 1 Hz and 1% strain. Hydrogel components were mixed immediately prior to measurement and added to the platter precooled to 4°C. With the geometry in place, the platter was rapidly warmed to 37°C and measurements began. It is estimated that 10–20 s passed between mixing components and measurements starting.
2.5. Scanning electron microscopy (SEM)
Hydrogels were produced as described, adding phosphate buffer (PB; 10 mM, 7.4 pH) instead of cell media to completely cover the hydrogels after cross-linking. Incubation at 37°C was continued for further 24 h. The resulting hydrogels were carefully placed onto carbon disc sample holders, rapidly frozen with liquid nitrogen and lyophilized. The dried samples were carefully sliced to expose the internal structure and then sputter-coated with platinum for 10 s. The SEM analysis was conducted with a LEO 1550 Gemini microscope (Zeiss, Germany) operated at a voltage of 3 kV.
2.6. SH-SY5Y cell culture
SH-SY5Y cells were obtained from ATCC, U.S.A (CRL-2266) and were thawed and cultured as per the supplier’s instructions in DMEM/F12 (1:1) cell media (Biowest, U.S.A), where DMEM stands for Dulbecco’s modified Eagle medium, and F12 for Ham’s F12 medium. This medium was supplemented with 10% fetal bovine serum (FBS; Biowest, U.S.A), 1% penicillin G/streptomycin (Biowest, U.S.A), and 1% non-essential amino acids (Biowest, U.S.A), hereby denoted as SH-SY5Y maintenance media. Cells were cultured in T75 flask and split 1:4 when confluence exceeded 80%. The passage number of the cells was never allowed to exceed 20 above the passage number at which the cells were supplied.
2.7. Viability of SH-SY5Y cells
Hydrogel components were prepared using SH-SY5Y maintenance media as the carrier solution. Confluent SH-SY5Y cultures were trypsinized (Trypsin 0.25% in PBS, Biowest, U.S.A) for 2 min at 37°C with gentle agitation to ensure a homogenous single-cell suspension and counted with trypan blue and a Bürker chamber. Counted cells were pelletized at 120 g for 6 min and then suspended in the (PEG-Az)8 component. Hydrogels were formed as described above, with a cell count of 105 per 50 µl hydrogel volume. Cells were cultured for up to 6 days at 37°C and 5% CO2. At time points of 1, 3 and 6 days, the media was removed, and the wells gently washed with PBS. AlamarBlueTM Cell Viability Reagent (AB) (Invitrogen, U.S.A) was used as the viability assay, with 10% added to cell media and placed in each well. Incubation followed for 2 h at 37°C and 5% CO2, after which the AB solution was moved to a new 96 well plate for the measurement of absorbance at 570 nm, and 600 nm (±5 nm) using a TECAN infinite M1000 Pro (Tecan, Switzerland). The absorbance measurements were calculated into percentage reduction as an indicator of cell viability using the calculation set out in the official protocol.
2.8. FPA and U87 cell culture
All reagents were purchased from Thermo Fisher Scientific, MA, U.S.A. unless stated otherwise. Human primary fetal astrocytes here, referred to as FPA (ScienCell, U.S.A), were maintained as per the supplier’s instructions. In brief, the cells were cultured in tissue culture-treated T75 flasks coated with 1× Attachment Factor protein in ScienCell astrocyte media (AM) with the supplements of 2% FBS, 1% astrocyte growth supplement (AGS), and 1% PenStrep (all ScienCell). When confluent, the cells were passaged at a ratio of 1:4, using TrypLE incubation for 4 min. Defined Trypsin Inhibitor (DTI) was used for TrypLE inhibition; the cells were spun down to a pellet at 200 g, after which they were resuspended in the supplemented AM. The U87 glioblastoma cells, previously published by Pontén and MacIntyre [Citation21] and obtained from Uppsala University, were maintained in tissue culture-treated T75 flasks in high glucose DMEM supplemented with 10% FBS and 1% PenStrep. When confluent, the cells were passaged at a ratio of 1:4 by a 4 min TrypLE incubation. The cells were then spun down to a pellet at 200 g, after which they were resuspended in the supplemented DMEM described above.
2.9. Encapsulation of FPA and U87 cells
Two days before the encapsulation of the cells into 3D hydrogels, both the FPA and the U87 were gradually adjusted to serum-free media. Serum-free media made from AM supplemented with 1% G5 supplement, 1% AGS, and 1% PenStrep was mixed 1:1 with respective maintenance media of each cell type, upon which the media was given to the cells. One day before encapsulation, the media of the FPA and the U87 were changed fully to the serum-free media. 1.6 million cells/ml of FPA and U87 were encapsulated in 1% HA:PEG hydrogels containing 1 mM of either peptide and carefully pipetted into half-area 96 well plates in gels of 50 µl each. The cells were cultured for 6 days and compared to 2D cultures on standard tissue culture plates. The respective peptides’ influence was studied with AB viability measurement after 1, 3, and 6 days of culture in the hydrogels. The AB viability assay was adapted to 3D cell culture format by increasing the incubation time of the AB reagent to 2 h before media sampling was performed. Longer incubation times were also evaluated but resulted in saturation of the assay. After 6 days of subculture, the cells were lysed, and mRNA expression of the intermediate filament protein Vimentin (VIM), the focal adhesion protein Paxillin (PAX), integrin subunit beta 1 (ITGB1), the focal adhesion kinase Protein Tyrosine Kinase 2 (PTK2), and the HA receptor CD44 were investigated for both cell types with qPCR. Glial fibrillary acidic protein (GFAP) and calcium-binding protein B (S100B) were additionally assayed for the FPA. The samples were studied as technical duplicates and contained the gene of interest (GOI) and the chosen housekeeping gene (HKG) GAPDH. The expression of the GOI is presented as normalized to the reference sample (RS), in this case, 2D-cell cultures, according to the following calculation:
Origin Pro was used to calculate p-values using Linear Mixed Models (LMM). Any samples measured to a Ct higher than 35 and technical duplicates with a variation higher than one were excluded.
Both cell types were stained for CD44, F-actin, and nuclei to visualize the cellular morphology and imaged with a confocal microscope. The confocal images were composed by maximum projection, after which brightness and contrast were adjusted to accentuate best the structures illustrating the cellular morphology. The image analysis was carried out in Fiji [Citation22]. To quantify the actin filament spreading and branchpoints the FIJI macro TWOMBLI was used with the following parameters: contrast saturation 0.35, line width 15, curvature window 50, minimum branch length 10, and minimum gap diameter 0 [Citation23].
2.10. 3D bioprinting
All 3D bioprinting was performed using a syringe extrusion printhead on a Cellink BIOX 3D bioprinter (BICO, U.S.A). FPA were cultured until confluent and then detached using TrypLE express. The cells were suspended in the (Peg-Az) 8 component at a concentration of 200 000 per 100 µl of a final hydrogel. Hydrogel components were prepared and combined as described above, with the exception that Cy5-azide was added at a concentration of approximately 40 µM to facilitate imaging of bioprinted structures. Hydrogel grid structures were printed directly on sterile tissue-culture treated plates, sealed, and incubated at 37°C and 5% CO2 for 1 hour for gelation to occur. After one-hour, pre-warmed co-culture media was added, and the plates were cultured for 4 days, after which the grid structures were fixated with 0.5% paraformaldehyde in preparation for staining.
3. Results and discussion
3.1. Hydrogel formation
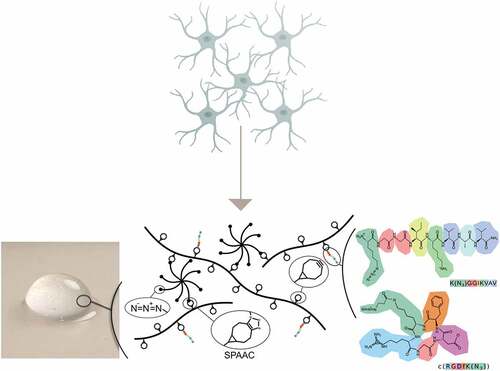
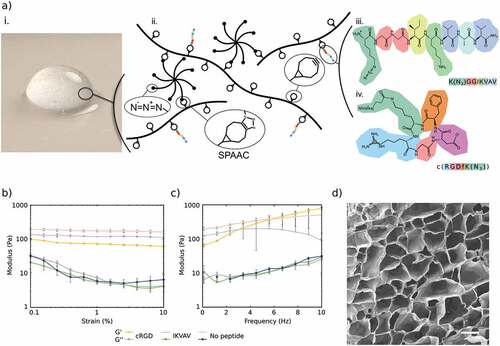
HA-based hydrogels were obtained using copper-free click-chemistry as described previously [Citation15,Citation24], using bicyclo[6.1.0]nonyne (BCN) modified HA (≈100 kDa) and an 8-arm PEG with terminal azide (Az) groups ((PEG-Az) 8) as a cross-linker (). The modularity of the hydrogel system allows for tuning of the viscoelastic properties, which were optimized for culture of neuronal and glial cells. The storage modulus was about 175 Pa at 1% strain and 1 Hz, which is within the typical range for neural tissue [Citation25]. Electron micrographs of the hydrogels () show non-fibrillar homogeneous hydrogels with a microporous structure as a result of ice crystal formation during freeze-drying prior to imaging. To promote cell–hydrogel interactions, an azide-functionalized cell adhesion peptide (cRGD or IKVAV) was conjugated to the BCN-modified HA prior to cross-linking. Due to the loss of some BCN groups, the peptide functionalized hydrogels showed a minor but not statistically significant decrease in the stiffness of the hydrogels (). This is similar to observations in prior work where functionalization of the hydrogels had limited effects on the viscoelastic properties [Citation26]
Figure 1. a) i. Photographic image of HA-based hydrogel. ii. schematic illustration of the HA:PEG: peptide system with the detailed amino acid structures of the peptides iii. IKVAV and iv. cRGD. b) Strain sweeps from 0.1 to 10% and at 1 Hz of hydrogels, and c) frequency sweeps from 0.1 to 10 Hz and at 1% strain of hydrogels with 1% (w/v) HA-BCN:(PEG-Az)8. Error bars represent standard deviation (n=3). d) SEM image of lyophilized 1% (w/v) HA-BCN:(PEG-Az)8 hydrogel. Scale bar is 50 µm.
3.2. SH-SY5Y viability
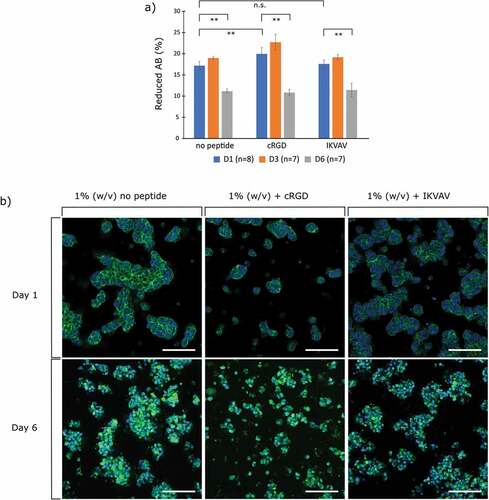
For initial evaluation of hydrogel biocompatibility, we employed the widely used SH-SY5Y neuroblastoma cells and assessed their viability using Alamar blue (AB) at time points of 1, 3 and 6 days (). The percentage of reduction of AB was used as an indicator for cell viability since it indicates metabolically active cell populations. At the day 1 time point, there was a significant increase in metabolic activity between the cRGD and no peptide condition; however, there was no significance between the IKVAV and the no peptide condition for the same time point. After 6 days in culture, all three conditions showed a significant decrease in metabolic activity. The 3D cell cultures were then fixated and stained for F-actin () at time points 1 and 6. The representative images show that the cells in the cRGD functionalized hydrogels were sparser but were equally less prone to form large spheroids or cell clusters. Such cell clustering is evident in the IKVAV-functionalized hydrogel and even more so in the no peptide condition. Clustering of cells is a known trait for the SH-SY5Y cell line and does not necessarily indicate a non-viable cell population [Citation27]. Furthermore, while a decrease in metabolic activity was observed between day 1 and day 6, the micrographs show an increase in cell number, indicating proliferation both without and with peptides. The cell clusters appeared less dense over time with more condensed cells, which has previously been observed for undifferentiated SH-SY5Y cultured on soft substrates, also accompanied by a decrease in cell stiffness indicating lower degrees of actin polymerization [Citation28,Citation29]. The hydrogels thus provide adequate and similar support for cell proliferation irrespectively of peptide functionalization, on par with observations of SH-SY5Y in other soft engineered hydrogels.
Figure 2. a) Cell viability of SH-SY5Y cell cultures using Alamar blue (AB). Values correspond to the percentage of AB reduced by viable cells on day 1 and 6. Error bars are standard deviation and significance is calculated using ANOVA with Tukey HSD (n.s. = no significance/>0.05), ** = <0.01). b) Micrographs of 3D cell cultures of SH-SY5Y cells in 1% (w/v) HA-BCN:(PEG-Az)8. Green is phalloidin staining for F-actin, blue is Hoechst staining for nucleus. Scale bar = 100 µm.
3.3. Viability of encapsulated FPA and U87 cells
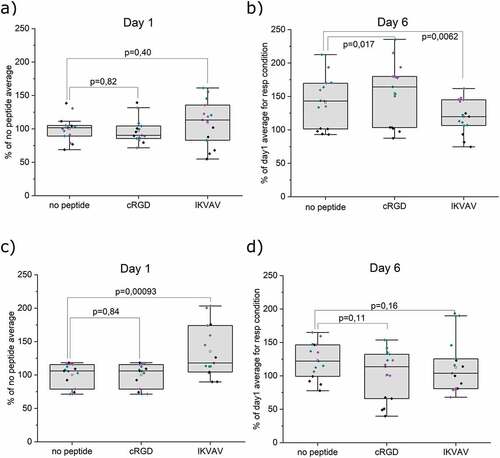
To assess the possibility to support astrocytic cells, primary human astrocytes were encapsulated in 3D hydrogels. Viability was studied using the AB viability assay. After 1 day of subculture, FPA show comparable viability in all gel conditions, . The addition of cRGD or IKVAV did not affect the viability at this stage. Over the course of 6 days (), all conditions lead to an increase in FPA viability compared to the corresponding hydrogel condition at 1 day of subculture. Moreover, a significant increase in viability was seen compared to the no-peptide HA when cRGD was added to the hydrogel. Surprisingly, the increase in viability from day 1 to day 6 was smaller with the addition of IKVAV. Regardless, these results indicate that the hydrogels are suitable for maintaining FPA. However, peptide functionalization had limited effect on promoting proliferation. These results align with our previous observations for the culture and differentiation of neural cells [Citation26]. In addition to the culture of primary astrocytes, we explored the often-used astrocytic model of the glioblastoma cell line U87. AB assay after 1 day of subculture shows viable cells in all gel conditions. The addition of IKVAV to the hydrogels resulted in a significant increase in initial U87 viability, while the addition of cRGD had no effect. The viability remained stable for all conditions after 6 days of culture, with no significant effects from the addition of cRGD or IKVAV. The effects of the hydrogels on the viability of U87 were thus similar to that of FPA, but in clear contrast to SH-SY5Y. With the neuroblastoma cells, cRGD rather than IKVAV improved short-term viability, and viability decreased significantly by day 6 for all conditions. These data suggest that the different cell lineages rely on distinct dominant mechanisms for interaction with the hydrogels.
Figure 3. AB viability assay of encapsulated cells. a) Viability of FPA after 1 day in respective hydrogel conditions. b) Change in viability of FPA after 6 days in respective hydrogels relative to day 1. c) Viability of U87 after 1 day in respective hydrogel conditions. d) Change in viability of U87 after 6 days in respective hydrogels relative to day 1. Data were collected from three individual experiments (indicated with different colors. N=5, where N represents a hydrogel replicate. Origin Pro was used for derivation of p-values with the LMM method, all data included. Data are presented both as a box indicating the 25th–75th percentile, including a median line and ±1.5 IQR whiskers, with the addition of individual data points.
3.4. mRNA expression analysis of encapsulated FPA and U87
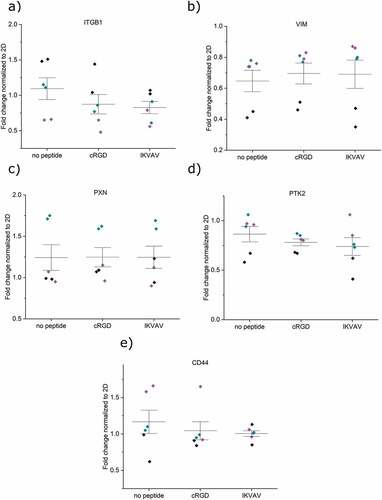
After 6 days of culture in the hydrogels, the cells were lysed, and mRNA expression levels were measured and normalized with respect to cells cultured in conventional 2D tissue culture plates. (). For FPA, five attachment- and two astrocytic-related markers were evaluated. For U87, the two astrocytic-related markers were left out in the analysis. Integrin ITGB1 was detected in both cell types, with no difference in expression for any of the hydrogel conditions or cell types () with p-values presented in Table S1 a) and b). The intermediate filament protein vimentin (VIM), the focal adhesion protein paxillin (PAX), and the focal adhesion kinase protein tyrosine kinase 2 (PTK2) showed no changes in expression between cells cultured in non-functionalized hydrogels and hydrogels functionalized with cRGD or IKVAV (), see Table S1 for p-values. These findings indicate that the cells continue to probe and interact with their pericellular microenvironment also in the absence of engineered integrin-binding motifs. Previous studies have found that cells encapsulated within non-adhesive HA-based hydrogels indeed both secrete fibronectin and express integrins as a result of reciprocal cell–hydrogel interactions, which can influence the cell fate [Citation30]. The maintained expression levels of integrins, PAX, and PTK2 in non-peptide functionalized hydrogels could thus likely be a result of cell-mediated formation of a proteinaceous pericellular matrix that would allow for integrin-mediated interactions. The role of integrin-mediated cell–hydrogel interactions was further underscored by the observed limited role of CD44. No differences in HA-binding CD44 expression were seen for any of the cell types and between the different hydrogel conditions (). Attempts to block the dimerization of CD44 have previously been observed to be an efficient way of controlling cellular attachment in tumors, suggesting that such studies can be interesting for understanding astrocyte adhesion to the HA:PEG hydrogels used here [Citation31,Citation32]. We thus further investigated CD44–hydrogel interactions by blocking the receptor with both antibodies and a small molecule-compound. Interestingly, the data did not indicate any major changes in cell–hydrogel interactions (data not shown). However, a larger panel of CD44 blocking methods would be needed to fully exclude this potential mode of cell–hydrogel interaction. Furthermore, two astrocytic markers were assayed for FPA; the calcium-binding protein B (S100B) and the glial fibrillary acidic protein (GFAP). No difference in mRNA expression levels was seen upon functionalization of the hydrogels with neither IKVAV nor cRGD (), indicating that astrocyte phenotype was not influenced by additional integrin-mediated interactions between FPA and the hydrogels. The expression of all studied genes thus remained conserved across the 2D and 3D culture systems studied here and suggests that we have a stable in vitro phenotype in both conditions.
Figure 4. mRNA expression analysis of encapsulated FPA after 6 days in different hydrogel conditions a) ITGB1, b) VIM, c) PXN, d) PTK2, e) CD44, f) S100B, and g) GFAP. All samples contain the housekeeping gene GAPDH. Origin Pro was used for the derivation of p-values with the LMM method. Data were collected as duplicates from three independent experiments, indicated with different colors. Whiskers indicate ±1 SE around the mean (bar).
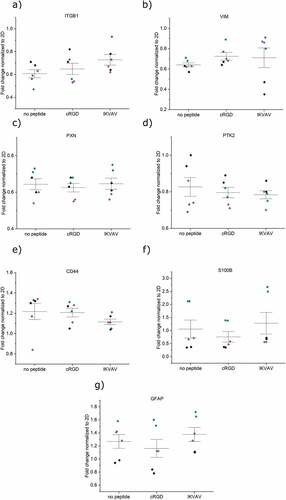
Figure 5. mRNA expression analysis of encapsulated U87 after 6 days in different hydrogel conditions a) ITGB1, b) VIM, c) PXN, d) PTK2, and e) CD44. All samples contain the housekeeping gene GAPDH. Origin Pro was used for the derivation of p-values with the LMM method. Data were collected as duplicates from three independent experiments, indicated with different colors. Whiskers indicate ±1 SE around the mean (bar).
3.5. Morphology of encapsulated FPA and U87 cells
The interactions between the cells and the hydrogels were further studied by confocal microscopy and immunostaining of the HA-binding CD44 receptor and F-actin staining for visualization of cytoskeletal organization (). Encapsulated FPA cultured in hydrogels without peptides appeared to primarily grow as single cells with a slightly condensed soma, while some cells spread out their protrusions and connected with neighboring cells and attached to the hydrogel (). CD44 was partially located as a halo surrounding the nuclei and was expressed along the soma and cellular protrusions. Similarly, F-actin staining revealed a rounded shape of a subpopulation of the cells, while other cells appeared more stellate and clearly interacting with the hydrogel matrix and other cells. Cell nuclei appeared oval and healthy under all conditions and only a small minority of the cells showed condensed nuclei with a bright and smaller appearance. The conjugation of cRGD or IKVAV to the hydrogel resulted in a slightly different morphology where cells extended their protrusions and spread out to a greater extent, seen as a larger surface area of the individual cells and more extensive cell–cell contacts. The localization of CD44 was similar in both the cRGD and IKVAV functionalized hydrogels and was seen both in the soma and along the cellular extensions. F-actin staining shows in detail how the cells spread out and connected to other cells nearby.
Figure 6. Confocal images of encapsulated FPA (a) and U87 (b) in respective hydrogels after 6 days. FPA and U87 are stained with HA receptor CD44 (magenta), cytoskeletal F-actin marker Phalloidin (cyan), and nuclear stain Hoechst (gray). 80 000 cells were seeded per hydrogel and for this analysis, six hydrogel replicates per condition were imaged at four sites per hydrogel. Scale bar: 30 μm.
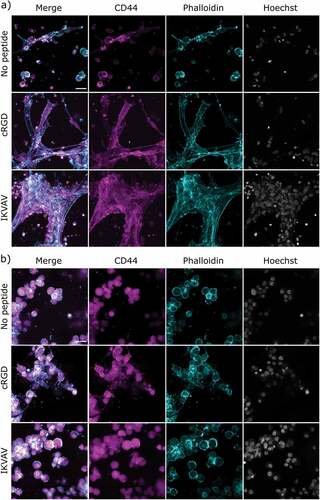
In contrast to the extensive spreading of FPA in the hydrogels, U87 cells typically appeared clustered and rounded; overall they appear very similar to SH-SY5Y. All hydrogel conditions resulted in cells positive for CD44. The localization was limited to an area surrounding the cell nuclei (). The only exception was seen for the cRGD condition, where small extended intercellular connections were observed between certain cells. Similarly, F-actin staining visualized the partially condensed soma and little or no visible protrusions or cell–cell connections, apart from in cRGD functionalized hydrogels where some connections between cells were observed. Nuclei for all hydrogel conditions appeared healthy, although with a rounder shape than what was observed for FPA cultured under identical conditions.
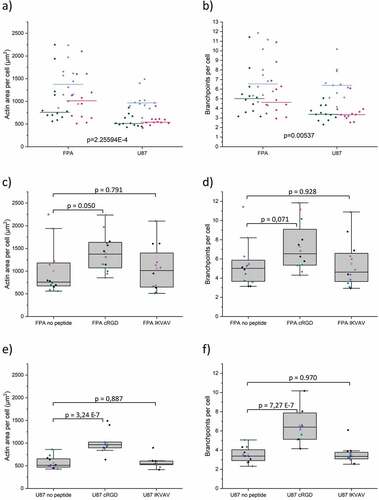
We further employed quantitative image analysis of the F-actin networks (in terms of area per cell, and branch points per cell) to expand on our qualitative analysis above. These data fully support the notion that FPA showed a more stellate morphology compared to U87 across all gel conditions (). In contrast to the observation from the qualitative image analysis, IKVAV functionalized hydrogels did not show any significant changes in these parameters for either cell type. This could be attributed to the fact that the observed FPA networks in IKVAV-functionalized hydrogels were also accompanied by larger cell numbers compared to cRGD-functionalized hydrogels. The data () do in fact support higher variability in cell morphology, which is also evident from and Figure S4. This correlates with the distinct cell viability changes observed in the presence of IKVAV (). Specifically, the higher variability of day 1 FPA survival, combined with lower proliferation toward day 6, would potentially support robust network formation in some cells, while a majority remain condensed. For U87 (), a comparison of morphology and viability data for cells in IKVAV-functionalized cells indicates that although the presence of IKVAV supported cell survival and proliferation, there was no increase in cell spreading and branching. The most notable finding from the analysis, however, is that in agreement with the qualitative assessment, the addition of cRGD successfully and significantly promoted spreading and branching of both cell types.
Figure 7. Quantification of actin filament spreading and branchpoints in FPA and U87 in respective hydrogels after 6 days. a) Actin area per cell across all gel conditions. b) Branchpoints per cell across all gel conditions. In a-b) green is the condition no peptide, blue is cRGD and magenta is IKVAV c) Actin area per FPA cell. d) Branchpoints per FPA cell. e) Actin area per U87 cell. f) Branchpoints per U87 cell. Data were collected from three individual experiments N=5, where N represents a hydrogel replicate. In c-f) the different colors represent the different individual experiments. Origin Pro was used for derivation of p-values with the LMM method, all data included. Data are presented both as a box indicating the 25th–75th percentile, including a median line and ±1.5 IQR whiskers, with the addition of individual data points.
3.6. 3D bioprinting
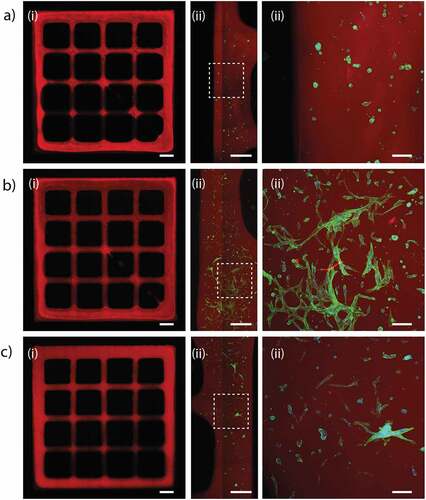
To finally investigate the possibilities of using FPA-laden hydrogels as a bioink for 3D bioprinting, we printed hydrogel structures in a grid pattern, followed by investigating cell number and morphology (). Despite being rather soft, the HA-based bioinks could be printed with good resolution. No apparent differences in cell density could be seen for the different hydrogel compositions. Cells printed in hydrogels without peptides grew as single rounded cells with a slightly condensed soma. Compared to hydrogel casting, the more condensed morphology observed after printing is likely a result from added stress from the bioprinting process. However, the presence of cell adhesion motifs resulted in cells with defined protrusions and multiple connections between neighboring cells in peptide-functionalized bioinks. With IKVAV, the difference compared to control appeared more distinct here than in cast hydrogels. Branching and spreading were, however, substantially more pronounced for FPA printed using the cRGD-functionalized hydrogels, which is in line with the observations from the conventional 3D cell culture. During the shorter time-lines and the shear stress induced by the bioprinting process, the availability of hydrogel-integrated cell-adhesion motifs thus appears to be more critical for FPA spreading compared to when cultured under longer times in 3D cast hydrogels.
Figure 8. Bioprinted FPA at day 4. The cells were encapsulated in HA-BCN:(PEG-Az)8 ± cRGD/IKVAV. a) HA-BCN:(PEG-Az)8. b) HA-BCN:(PEG-Az)8 functionalized with cRGD. c) HA-BCN:(PEG-Az)8 functionalized with IKVAV. The HA backbone was labelled with Cy5-Az (red), and FPA were stained green with phalloidin for F-actin. i) Full grid structure, scale bars are 1 mm. ii) Zoom in on part of the structure, scale bars are 0.5 mm. iii) Zoomed in view as indicated with white rectangles, scale bars are 100 µm. The images are representative of a larger set of imaged grid structures (N = 4–8).
4. Conclusions
This work presents an evaluation of the 3D culture of cell models SH-SY5Y and U87 glioma cells and human FPA in a modular and bioprintable HA-based hydrogel system. The cells were cultured over a 6-day period. All three cell models expressed the HA binding receptor CD44 but displayed distinctly different morphologies. FPA were seen to spread out their processes and connect to neighboring cells in a network-forming pattern, especially in hydrogels functionalized with cRGD-peptides. In contrast, U87 and SH-SY5Y cells mainly formed clusters of cells showing little or no large-scale network formation under any of the conditions. mRNA expression analysis of attachment and astrocytic markers showed no differences between cells cultured within hydrogels without integrated adhesion motifs and hydrogels functionalized with IKVAV or cRGD. This was likely as a result of the formation of a cell-secreted pericellular proteinaceous matrix that could mediate integrin-interactions with their microenvironment. Bioprinting of FPA resulted in hydrogel-dependent morphologies. Bioinks comprising hydrogels without peptide-cell adhesion motifs appeared rounded, whereas cells in hydrogels modified with IKVAV and especially cRGD showed branched morphologies with extensive cell–cell contacts 4 days post printing. These engineered hydrogels enable the possibilities to bioprint, culture and maintain FPA and can thus facilitate the development of more elaborate neural and astrocytic tissue and disease models.
Acknowledgments
I.M. and M.J. contributed equally to this work. The financial support from the Swedish Foundation for Strategic Research (SFF FFL15-0026) and the Knut and Alice Wallenberg Foundation (KAW 2016.0231,2021.0186) is gratefully acknowledged.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Allen NJ, Bennett ML, Foo LC, et al. Astrocyte glypicans 4 and 6 promote formation of excitatory synapses via GluA1 AMPA receptors. Nature. 2012;486(7403):410–15.
- Tsai Y-J, Huang C-T, Lin S-C, et al. Effects of regional and whole-body hypothermic treatment before and after median nerve injury on neuropathic pain and glial activation in rat cuneate nucleus. Anesthesiology. 2012;116(2):415–431.
- Barres BA. The mystery and magic of glia: a perspective on their roles in health and disease. Neuron. 2008;60(3):430–440.
- Hawkins BT, Davis TP. The blood-brain barrier/neurovascular unit in health and disease. Pharmacol Rev. 2005;57(2):173–185.
- Abbott NJ, Rönnbäck L, Hansson E. Astrocyte–endothelial interactions at the blood–brain barrier. Nat Rev Neurosci. 2006;7(1):41–53.
- Thomsen MS, Routhe LJ, Moos T. The vascular basement membrane in the healthy and pathological brain. J Cereb Blood Flow Metab. 2017;37(10):3300–3317.
- Johnson KM, Milner R, Crocker SJ. Extracellular matrix composition determines astrocyte responses to mechanical and inflammatory stimuli. Neurosci Lett. 2015;600:104–109.
- Yudkoff M, Pleasure D, Cregar L, et al. Glutathione turnover in cultured astrocytes: studies with [15N] glutamate. J Neurochem. 1990;55(1):137–145.
- Sagara J, Makino N, Bannai S. Glutathione efflux from cultured astrocytes. J Neurochem. 1996;66(5):1876–1881.
- Huang S-F, Othman A, Koshkin A, et al. Astrocyte glutathione maintains endothelial barrier stability. Redox Biol. 2020;34:101576.
- Oberheim NA, Tian G-F, Han X, et al. Loss of astrocytic domain organization in the epileptic brain. J Neurosci. 2008;28(13):3264–3276.
- Schiweck J, Eickholt BJ, Murk K. Important shapeshifter: mechanisms allowing astrocytes to respond to the changing nervous system during development, injury and disease. Front Cell Neurosci. 2018;12.
- Rocha DN, Ferraz-Nogueira JP, Barrias CC, et al. Extracellular environment contribution to astrogliosis—lessons learned from a tissue engineered 3D model of the glial scar. Front Cell Neurosci. 2015;9.
- Huettner N, Dargaville TR, Forget A. Discovering cell-adhesion peptides in tissue engineering: beyond RGD. Trends Biotechnol. 2018;36(4):372–383.
- Christoffersson J, Aronsson C, Jury M, et al. Fabrication of modular hyaluronan-PEG hydrogels to support 3D cultures of hepatocytes in a perfused liver-on-a-chip device. Biofabrication. 2018;11(1):015013.
- Lam J, Carmichael ST, Lowry WE, et al. Hydrogel design of experiments methodology to optimize hydrogel for iPSC-NPC culture. Adv Healthc Mater. 2015;4(4):534–539.
- Tatman PD, Muhonen EG, Wickers ST, et al. Self-Assembling peptides for stem cell and tissue engineering. Biomater Sci. 2016;4(4):543–554.
- Cheng T-Y, Chen M-H, Chang W-H, et al. Neural stem cells encapsulated in a functionalized self-assembling peptide hydrogel for brain tissue engineering. Biomaterials. 2013;34(8):2005–2016.
- Kim Y, Kumar S. CD44-Mediated adhesion to hyaluronic acid contributes to mechanosensing and invasive motility. Mol Cancer Res. 2014;12(10):1416–1429.
- Selegård R, Aronsson C, Brommesson C, et al. Folding driven self-assembly of a stimuli-responsive peptide-hyaluronan hybrid hydrogel. Sci Rep. 2017;7:7013.
- Pontén J, Macintyre EH. Long term culture of normal and neoplastic human glia. Acta Pathologica Microbiologica Scand. 1968;74(4):465–486.
- Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682.
- Wershof E, Park D, Barry DJ, et al. A FIJI macro for quantifying pattern in extracellular matrix. Life Sci Alliance. 2021;4(3).
- Aronsson C, Jury M, Naeimipour S, et al. Dynamic peptide-folding mediated biofunctionalization and modulation of hydrogels for 4D bioprinting. Biofabrication. 2020;12(3):035031.
- Budday S, Ovaert TC, Holzapfel GA, et al. Fifty shades of brain: a review on the mechanical testing and modeling of brain tissue. Arch Computat Methods Eng. 2020;27(4):1187–1230.
- Jury M, Matthiesen I, Rasti Boroojeni F, et al. Bioorthogonally cross-linked hyaluronan–laminin hydrogels for 3D neuronal cell culture and biofabrication. Adv Healthcare Mater. 2022;11:2102097.
- Kovalevich J, Langford D. Considerations for the use of SH-SY5Y neuroblastoma cells in neurobiology. In: Amini S White M, editors. Neuronal cell culture: methods and protocols. Totowa(NJ): Humana Press; 2013. p. 9–21.
- Ozgun A, Erkoc-Biradlı FZ, Bulut O, et al. Substrate stiffness effects on SH-SY5Y: the dichotomy of morphology and neuronal behavior. J Biomed Mater Res, Part B. 2021;109(1):92–101.
- Kruger TM, Bell KJ, Lansakara TI, et al. Reduced extracellular matrix stiffness prompts SH-SY5Y cell softening and actin turnover to selectively increase Aβ(1–42) endocytosis. ACS Chem Neurosci. 2019;10(3):1284–1293.
- Ferreira SA, Motwani MS, Faull PA, et al. Bi-directional cell-pericellular matrix interactions direct stem cell fate. Nat Commun. 2018;9(1):4049.
- Hei B, Wang J, Wu G, et al. Verbascoside suppresses the migration and invasion of human glioblastoma cells via targeting c-met-mediated epithelial-mesenchymal transition. Biochem Biophys Res Commun. 2019;514(4):1270–1277.
- Wang C, Wang Z, Chen C, et al. A low MW inhibitor of CD44 dimerization for the treatment of glioblastoma. Br J Pharmacol. 2020;177(13):3009–3023.