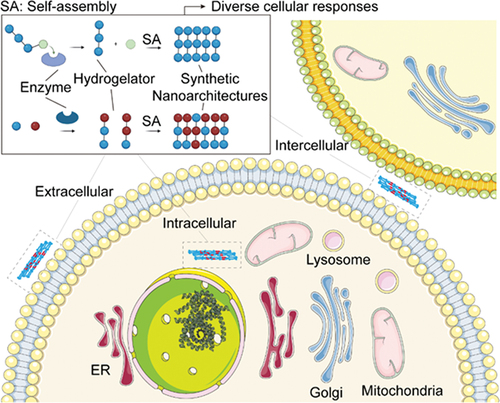
ABSTRACT
Nanoarchitectonics, as a technology to arrange nano-sized structural units such as molecules in a desired configuration, requires nano-organization, which usually relies on intermolecular interactions. This review briefly introduces the development of using enzymatic reactions to control intermolecular interactions for generating artificial nanoarchitectures in a cellular environment. We begin the discussion with the early examples and uniqueness of enzymatically controlled self-assembly. Then, we describe examples of generating intracellular nanostructures and their relevant applications. Subsequently, we discuss cases of forming nanostructures on the cell surface via enzymatic reactions. Following that, we highlight the use of enzymatic reactions for creating intercellular nanostructures. Finally, we provide a summary and outlook on the promises and future direction of this strategy. Our aim is to give an updated introduction to the use of enzymatic reaction in regulating intermolecular interactions, a phenomenon ubiquitous in biology but relatively less explored by chemists and materials scientists. Our goal is to stimulate new developments in this simple and versatile approach for addressing societal needs.
GRAPHICAL ABSTRACT
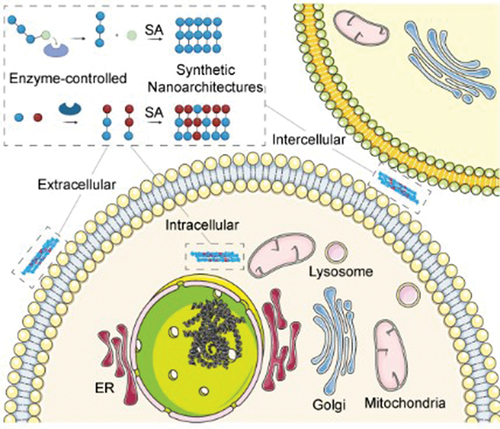
IMPACT STATEMENT
Enzymatic reactions in cells create precise nanoarchitectures, offering insights into cell biology through controllable nanoarchitectonics, as shown by numerous examples in this review.
Introduction
Nanoarchitectonics is a concept that integrates nanotechnology methodologies with diverse research disciplines, particularly supramolecular chemistry, to harness self-assembly processes crucial in various biological systems, such as cells. The driving force behind building supramolecular assemblies is noncovalent interaction [Citation1]. Thus, modulating intermolecular, noncovalent interactions is essential for generating supramolecular assemblies. In this regard, nature has devised ingenious designs to control noncovalent interactions, evident in the evolution of life. Cells, for instance, contain numerous nanoarchitectonics held together by noncovalent interactions [Citation2]. These structures dynamically interact with each other to carry out fundamental biological functions.
Enzymatic reactions serve as the fundamental mechanism for controlling intermolecular interactions inside cells. For example, multiple enzymatic reactions create lipids [Citation3], the building blocks that form cell membranes to provide compartmentalization. Cytoskeletons, such as actin filaments or microtubules, are maintained by enzymatic dephosphorylation [Citation4]. The dynamic characteristics of the endoplasmic reticulum membrane involve enzymatic reactions to regulate network formation with proteins and phospholipids [Citation5]. Inflammasomes, acting as central organization centers for cellular host defense, are created through the self-assembly of proteins and governed by enzymatic posttranslational modifications [Citation6–8]. These findings emphasize the significance of enzymatic reactions in orchestrating noncovalent interactions for non-equilibrium self-assembly that gives rise to supramolecular assemblies with biological functionalities.
The utilization of enzyme reactions and self-assembly by nature to maintain life has inspired the integration of enzymatic reactions and self-assembly for generating nanoarchitectonics. While the early focus of enzymatic formed nanostructures center on soft materials, such as supramolecular hydrogels [Citation9,Citation10], the development of a multistep process known enzyme-instructed self-assembly (EISA) [Citation11] has allowed the creation of nanostructures in cellular environment for increasing numbers of applications over the last two decades. This review aims to offer a concise review on the use of enzymatic reactions to regulate intermolecular interactions for generating artificial nanoarchitectures within the cellular environment (). We start with an examination of early examples and the discussion of the uniqueness of enzymatically controlled self-assembly processes. Subsequently, we describe the use of enzymatic reactions for generating intracellular nanostructures and highlight the applications that stem from these intricate nanostructures within the cellular milieu. Following that, we discuss enzymatically the formation of nanostructures on the cell surface, particularly for profiling enzyme activities on cell surface. Then, we present intercellular nanostructures facilitated by enzymatic reactions, especially in the context of transcytosis and cell morphogenesis. Finally, we summarize the results presented and offer an outlook of this promising strategy. By providing an overview of applications of EISA within the cellular environment, we hope to illustrate the potential of the strategy and provide a starting point for molecular scientists to explore enzymatic reactions and self-assembly for generating nanoarchitectures with emergent properties for various applications.
Uniqueness of enzymatically controlled self-assembly
The prerequisite to build artificial nanostructures in cells is that the self-assembling building block needs to be amphiphilic, as the case of the lipids to form cell membranes. The theoretical studies of the intermolecular interactions of lipids or surfactants have already established the thermodynamic principle for self-assemblies of amphiphilic molecules. A key concept to correlate molecular structures with self-assembling behaviors is molecular packing parameter, providing rationalization and sometimes predicting molecular self-assembly in surfactant solutions. The molecular packing parameter, denoted as v0/a0l0, is expressed in terms of the surfactant tail volume (v0), tail length (l0), and the head group area per molecule (a0) at the aggregate surface. Through straightforward geometric relationships, a specific shape and size for the equilibrium aggregate can be derived from a given value of the molecular packing parameter [Citation12]. For example, when v0/a0l0 <1/3, the surfactants self-assemble to form micelles, and when 1/3 < v0/a0l0 <1/2, the micelles would transform to worm-like micelles.
Obviously, it is possible to use enzymatic reactions to reduce the head group area, thus increasing of packing parameter and promoting a change in micelles to a worm micelle shape. On the other hand, it is possible to use enzymatic reaction to increase the volume of the hydrophobic tails, thereby increasing the molecular packing parameter for the micelle-to-worm transition. Both cases have been realized by enzymatic reactions ().
Figure 1. Two modes of enzymatic self-assembly of molecules in water (hydrogelators); SA means self-assembly. (a) Use of a phosphatase to break O-P bond to produce hydrogelators that form supramolecular nanofibers in water. (b) Use of thermolysin to catalyze bond formation for producing hydrogelators that self-assemble in water to form nanofibers.
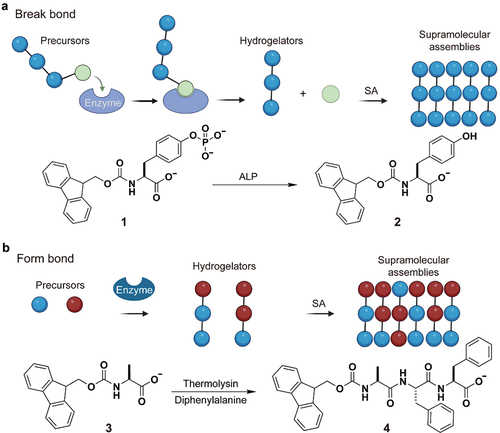
In the former case, an enzymatic reaction breaks a bond to remove hydrophilic phosphates group from amphiphilic fluorenylmethoxycarbonyl (Fmoc)-tyrosine phosphates (1), forming Fmoc-tyrosine (2), which self-assembles into nanofibers to form hydrogel () [Citation13]. In the latter case, an enzymatic reaction forms a bond to connect Fmoc-alanine (3) to diphenylalanine to form Fmoc-Ala-Phe-Phe (4), which self-assemble to form nanofibers that entangle to result in a hydrogel () [Citation14]. These concepts allow for the controlled assembly of molecular structures at interfaces, facilitated by an enzyme-driven process and a strategically designed seed layer [Citation15–19].
Interestingly, enzymes exhibit the ability to regulate their substrate to produce defect-free supramolecular nanofibers [Citation20], which appears to be a unique feature of this process that was termed as EISA [Citation11]. Another noteworthy and distinctive aspect of EISA lies in its capacity for self-assembly within conditions far from equilibrium. This occurs due to the impact of enzymatic reactions on the concentration of the self-assembling building blocks, which rarely stabilizes at equilibrium, although it may establish a steady state during the assembly process. Moreover, EISA, if occurs rapidly, can produce non-diffusive assemblies. This feature enables the targeted localization of supramolecular assemblies within a cellular environment, as elaborated in the subsequent sections.
Intracellular nanostructures
The enzymatic dephosphorylation utilized to initiate the self-assembly of simple small molecule in water offers a powerful strategy for constructing nanostructures inside cells. For example, in the earliest study [Citation21], enzymatic hydrogelation was achieved with E. coli, and the response of the E. coli was examined to illustrate this concept (). The soluble precursor (5) of a hydrogelator comprises a Phe-Phe motif, which is prone to self-assemble in water () [Citation22,Citation23], as the peptide backbone, a naphthylacetic acid as the N-terminal capping group, and a tyrosine phosphate as a C-terminal of 5 [Citation24]. After confirming that alkaline phosphatase (ALP) converts the solution of 5 into the hydrogel of 6 and that the nanofibers of 6 act as the network of the hydrogel, the authors used 5 to treat two types of E. coli strains-one overexpressing phosphatase and the other not. The overexpressed phosphatase in E. coli was found to catalyze the formation of the hydrogelator inside the bacteria (), and the subsequent intracellular hydrogelation inhibited the bacteria growth. This work, therefore, establishes the use of enzyme-instructed intracellular self-assembly of small molecules for creating artificial nanostructures and thus controlling the fate of cells.
Figure 2. (a) Schematic representation of intracellular nanofiber formation by enzymatic reactions. (b) Chemical structures of compound 5 and 6. (c) Concentration of 5 and 6 in culture media or inside bacteria. (a)-(c) are reproduced by permission from [Citation21], copyright [2007, John Wiley and Sons]. (d) Chemical structures of compound 7 and 8. (e) 3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide (MTT) assay of incubation of HeLa cells with esterase substrate 7. (d)-(e) are reproduced by permission from [Citation25], copyright [2007, John Wiley and Sons].
![Figure 2. (a) Schematic representation of intracellular nanofiber formation by enzymatic reactions. (b) Chemical structures of compound 5 and 6. (c) Concentration of 5 and 6 in culture media or inside bacteria. (a)-(c) are reproduced by permission from [Citation21], copyright [2007, John Wiley and Sons]. (d) Chemical structures of compound 7 and 8. (e) 3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide (MTT) assay of incubation of HeLa cells with esterase substrate 7. (d)-(e) are reproduced by permission from [Citation25], copyright [2007, John Wiley and Sons].](/cms/asset/f5e0ebc8-2be0-41c1-8407-5b7e4b46e07c/tsta_a_2373045_f0002_oc.jpg)
The cell selectivity of 5 against the phosphatase overexpressing E. coli implies that it is feasible to use enzymatic reactions to generated intracellular nanostructures for preferential inhibition of cancer cells while sparing normal cells. This assumption was confirmed by an esterase substrate (7) [Citation25] that differs from 5 only at the C-terminal. Specifically, replacing tyrosine phosphate in 5 by a 4-(2-aminoethoxy)-succinic acid affords 7 (). Carboxylesterase (CES) can remove the hydrophilic succinic acid from 7 to generate 8, a hydrogelator that self-assembles in an aqueous environment to form nanofibers. The onset of hydrogelation appears to be quite fast, occurring within 10 min under physiological condition and catalyzed by an esterase in aqueous buffer solution. When 7 was used to incubate with cancer cells (HeLa) or normal cells (NIH3T3), it only inhibits the growth of the cancer cells (), which expresses a higher level of esterases than the normal cells do. Further examination suggests that esterase in HeLa cells catalyze the formation of the hydrogelator, resulting in the formation of nanofibers as well as hydrogelation inside the HeLa cells, leading to cell death. However, the low-level expression of esterase in NIH3T3 cell is unable to generate enough hydrogelator to promote gelation, rendering the precursor innocuous to the NIH3T3 cells. This result provides an experimental foundation to use enzyme-instructed self-assembly (EISA) for targeting cancer cells [Citation25].
Since the demonstration of the concept of enzymatic formation of nanostructures inside cells, considerable of progresses have been made by multiple labs. For example, Rao et al. demonstrated the use of enzymatic cleavage to trigger rapid biorthogonal reactions for intracellular oligomerization, and the oligomers form nanoaggregates [Citation26]. Yang et al. have conjugated chemotherapy drug molecules to enzyme responsive peptides for intracellular drug delivery [Citation27]. Wang et al. have shown the application of enzymatic triggered assembly of drug-peptide conjugates in the tumors in mice models as an effective strategy for photodynamic therapy or chemotherapy [Citation28]. Ye et al. combined enzymatic reactions and multimodal imaging techniques to enable enhanced imaging of malignant tumors [Citation29]. Despite these advancements, the unambiguous demonstration of intracellular nanostructures has been elusive until 2020 [Citation30]. As shown in , it took a rather unusual peptide derivative (9) to generate nanofibers of 10 in both cell-free condition and inside cells (). The molecule 9 consists of a fluorophore (nitrobenzoxadiazole (NBD)), a self-assembling D-peptide backbone (D-Phe-D-Phe) [Citation22], a D-phosphotyrosine, and a C-terminal-L-lysine that is trimethylated (). The heterochirality and trimethylation appear to be key for the intracellular formation of nanofiber bundles because failing to satisfy either of these two requirements abolishes intracellular bundling, though the nanofibers still form in cell-free condition after the enzymatic phosphorylation of 9 to generate 10. The use of cryo-EM Iterative Helical Real Space Reconstruction (IHRSR) [Citation31] helical reconstructions reveals that 10 self-assembles into two distinct types of polymorphs, with one cross-structures possessing C7 symmetry and the other C2 symmetry. Molecular dynamics simulations suggest that the central pore of the filament contains water and ions to provide stabilization to the filament structure, which holds the NBD in the innermost layer. According to the Electron Tomography (ET), after Saos2 cell was incubated with 9 for 24 h, ALP inside the Saos-2 converts 9 to 10, which self-assembles to form monodispersed nanofibers that pack as twist bundles and extend from the plasma membrane to nuclear membrane. Fluorescent imaging indicates that endogenous cytoskeletons hardly interacting with the filaments of 10, although the filament bundles appear to be cytoskeleton-like (). While the macromolecular crowding inside cells likely enables the bundling of the filaments of 10, it remains unclear why heterochirality and trimethylation of the L-lysine are necessary for the intracellular bundling. Nevertheless, the work provides the first unambiguous example of intracellular nanoarchitecture form by synthetic molecules.
Figure 3. (a) Schematic illustration of intracellular enzymatic reaction to convert nanoparticles into filament bundles. (b) Chemical structures of compound 9 and 10. (c) Confocal laser scanning microscopy (CLSM) images of Saos-2 cells treated with 9 (200 mM) for different time periods. Scale bar = 10 μm. (a)-(c) are reproduced by permission from [Citation30], copyright [2020, Elsevier]. (d) Schematic illustration of ALP-instructed assembly for inhibiting cancer cells. (e) Chemical structures of compound 11 and 12. (f) IC50 values of 11 against Saos-2 or HepG2 cells. (g) Tumor volume of orthotopic osteosarcoma model established by Saos2-luc and Saos2-lung cells after tail intravenous injection of compound 11 or saline for 4 weeks. (h) Survival curves for orthotopic osteosarcoma nude mice (n = 5) treated with 11. (d)-(h) are reproduced by permission from [Citation32], copyright [2019, Elsevier].
![Figure 3. (a) Schematic illustration of intracellular enzymatic reaction to convert nanoparticles into filament bundles. (b) Chemical structures of compound 9 and 10. (c) Confocal laser scanning microscopy (CLSM) images of Saos-2 cells treated with 9 (200 mM) for different time periods. Scale bar = 10 μm. (a)-(c) are reproduced by permission from [Citation30], copyright [2020, Elsevier]. (d) Schematic illustration of ALP-instructed assembly for inhibiting cancer cells. (e) Chemical structures of compound 11 and 12. (f) IC50 values of 11 against Saos-2 or HepG2 cells. (g) Tumor volume of orthotopic osteosarcoma model established by Saos2-luc and Saos2-lung cells after tail intravenous injection of compound 11 or saline for 4 weeks. (h) Survival curves for orthotopic osteosarcoma nude mice (n = 5) treated with 11. (d)-(h) are reproduced by permission from [Citation32], copyright [2019, Elsevier].](/cms/asset/2b4be59a-0358-4f80-bc33-2fd9dd9515d5/tsta_a_2373045_f0003_oc.jpg)
Although the use of EISA is able to inhibit tumor effectively when the enzyme-responsive peptide conjugates with a chemotherapy drug, the question whether intracellular nanostructures alone can potently inhibit tumor growth remains an unanswered until a recent study to demonstrate that EISA of a phosphopeptide derivative (11) is able to effectively inhibit immunosuppressive tumor growth in mice models () [Citation32]. Specifically, consisting of a self-assembling peptide backbone (Nap-ff [Citation33]), a D-phosphotyrosine (yp), and a dimethyl-D-glutamate (eMe2), 11 self-assembles to form nanoparticles. ALP dephosphorylates 11 to form Nap-ffyeMe2 (12) (), which self-assembles to form rigid nanotubes with a diameter of 12 ± 2 nm. 11 is particularly potent for inhibiting immunosuppressive metastatic osteosarcoma, such as Saos2, because Saos2 overexpresses ALP, which converts ATP to an immunosuppressive signaling molecule, adenosine. Being converted to 12 by overexpressed ALP from Saos2, 11 results in Saos2 cell death within half an hour. Containing the dimethyl-D-glutamate group, 11 exhibits excellent cell selectivity, inhibiting Saos2 with IC50 of 4 µM, but its IC50 on the hepatocyte cells (HepG2) is over two orders of magnitude higher than the IC50 on Saos-2 (). Using an orthotopic murine model of osteosarcoma, in which the study shows that tail intravenous injection of 11 results in the 25-fold reduction in tumor volume of Saos2-lung, a Saos2 line metastasized to the lung (). As expected, the treatment of 11 significantly prolongs the survival time of the osteosarcoma-bearing nude mice (). Further examination also confirmed that 11 exhibits excellent biocompatibility to normal tissues and organs.
A rather unexpected benefit of using enzyme to instruct the formation of intracellular nanostructures is that it is feasible to generate the nanostructures with high spatial cellular resolution. That is, the substrates of enzymes can be used to target organelles in a cell-selective manner, such as targeting mitochondria, lysosome, endoplasmic reticulum (ER), and Golgi apparatus of cancer cells [Citation34–48]. For example, it is well established that mitochondria play essential roles in apoptosis and metabolism. Conventional approaches for targeting mitochondria rely on lipophilic and cationic molecules [Citation49,Citation50]. Although these lipophilic cations, when their concentrations are low, can precisely target mitochondria, they still lack cell selectivity.
In a study on the enzymatic supramolecular hydrogelation of branched peptides, it was serendipitously found that EISA of branched peptides can target mitochondria of cancer cells. Specifically, 13, consisting of a Nap-ffk as the backbone, NBD at the C-terminal, and the FLAG-tag (DYKDDDDK) [Citation51] as the side chain, is a substrate of enterokinase (ENTK) () [Citation35]. 13 self-assembles to form micelles, which turn into nanofibers upon enzymatic cleavage of the hydrophilic branch that contains FLAG motif (DDDDK) (). Fluorescent imaging suggests that the enzyme cleaved product (14) mainly localizes at mitochondria (). This mitochondrial localization is cell-selective and mostly occurs in cancer cells, which is probably due to a higher mitochondria membrane potential in cancer cells [Citation52]. This work illustrates a fundamentally new enzymatic process for targeting mitochondria by forming nanofibers selective on mitochondria.
Figure 4. (a) Chemical structures of compound 13 and 14. (b) Schematic illustration of ENTK instructed enzymatic reaction to convert micelles to nanofibers on mitochondria. (c) CLSM images of 13 and Mito-tracker in HeLa cells. Scale bar = 30 μm. (a)-(c) are reproduced by permission from [Citation35], copyright [2018, American Chemical Society]. (d) Chemical structures of compound 15 and 16. (e) Time-dependent CLSM images of 16 selectively form on HeLa cells and in HS-5 cells in the co-culture of HeLa and HS-5 cells ([Citation15] = 500 μM). Scale bar = 10 μm. (d)-(e) are reproduced by permission from [Citation54], copyright [2016, Elsevier]. (f) Chemical structures of compound 17 and 18. (g) Schematic illustration of the lysosome specific construction of nanofibers in living cells through multistage processes. (h) CLSM images of Saos-2 cells incubated with 17 (100 μM) for 4 h. Scale bar = 5 μm. (f)-(h) are reproduced by permission from [Citation41], [2021, John Wiley and Sons]. (i) Chemical structures of compound 19 and 20, as well as the TEM images of morphological transformation of 19 (0.5 wt %) after adding ALP (1 U/mL) into crescent-shaped aggregates. Scale bar = 50 nm. Reproduced by permission from [Citation42], [2018, American Chemical Society]. (j) Chemical structures of compound 21 and 22 and 23, and the involving enzymatic reactions. (k) Schematic illustration of the cellular uptake of 21 or 23 that is enzymatically processed into 22 to accumulate at Golgi; CMC means critical micelle concentration. (l) TEM images of 21 (5 μM) before and after the addition of ALP (0.1 U/mL) for 24 h. Scale bar = 100 nm. (j)-(l) are reproduced by permission from [Citation44], [2021, John Wiley and Sons]. (m) CLSM images of Galactose-1-phosphate uridylyltransferase red fluorescent protein (GalT-RFP) transfected HeLa cells treated with 21 (10 μM) for 8 minutes. Scale bar = 20 μm. (n) CLSM images of different cells (HeLa, Saos-2, HEK293, MCF-7, HS-5, SJSA-1, HepG2, B16F10, and S2) treated with 23 (10 μM) for 8 min. Scale bar = 20 μm.(m)-(n) are reproduced by permission from [Citation45], [2022, American Chemical Society].
![Figure 4. (a) Chemical structures of compound 13 and 14. (b) Schematic illustration of ENTK instructed enzymatic reaction to convert micelles to nanofibers on mitochondria. (c) CLSM images of 13 and Mito-tracker in HeLa cells. Scale bar = 30 μm. (a)-(c) are reproduced by permission from [Citation35], copyright [2018, American Chemical Society]. (d) Chemical structures of compound 15 and 16. (e) Time-dependent CLSM images of 16 selectively form on HeLa cells and in HS-5 cells in the co-culture of HeLa and HS-5 cells ([Citation15] = 500 μM). Scale bar = 10 μm. (d)-(e) are reproduced by permission from [Citation54], copyright [2016, Elsevier]. (f) Chemical structures of compound 17 and 18. (g) Schematic illustration of the lysosome specific construction of nanofibers in living cells through multistage processes. (h) CLSM images of Saos-2 cells incubated with 17 (100 μM) for 4 h. Scale bar = 5 μm. (f)-(h) are reproduced by permission from [Citation41], [2021, John Wiley and Sons]. (i) Chemical structures of compound 19 and 20, as well as the TEM images of morphological transformation of 19 (0.5 wt %) after adding ALP (1 U/mL) into crescent-shaped aggregates. Scale bar = 50 nm. Reproduced by permission from [Citation42], [2018, American Chemical Society]. (j) Chemical structures of compound 21 and 22 and 23, and the involving enzymatic reactions. (k) Schematic illustration of the cellular uptake of 21 or 23 that is enzymatically processed into 22 to accumulate at Golgi; CMC means critical micelle concentration. (l) TEM images of 21 (5 μM) before and after the addition of ALP (0.1 U/mL) for 24 h. Scale bar = 100 nm. (j)-(l) are reproduced by permission from [Citation44], [2021, John Wiley and Sons]. (m) CLSM images of Galactose-1-phosphate uridylyltransferase red fluorescent protein (GalT-RFP) transfected HeLa cells treated with 21 (10 μM) for 8 minutes. Scale bar = 20 μm. (n) CLSM images of different cells (HeLa, Saos-2, HEK293, MCF-7, HS-5, SJSA-1, HepG2, B16F10, and S2) treated with 23 (10 μM) for 8 min. Scale bar = 20 μm.(m)-(n) are reproduced by permission from [Citation45], [2022, American Chemical Society].](/cms/asset/df35d0cf-0daa-4657-a601-2e0bc1a3d084/tsta_a_2373045_f0004_oc.jpg)
Lysosomes, containing many hydrolytic enzymes for biomolecular degradation within cells, play essential roles in various physiological functions [Citation53]. Thus, considerable efforts have focused on selectively targeting lysosomes of desired cells. Since lysosomes are acidic and contain acid phosphatases (ACP), it is feasible to target lysosomes in a cell-selective and context-dependent manner. For example, A D-peptide derivative (15), containing a naphthylalanine and D-phosphotyrosine, can act as the substrate of either ALP or ACP () [Citation54]. After being dephosphorylated, 15 transforms into 16, which self-assembles to form nanofibers. In the co-culture of cancer (HeLa) and bone marrow stromal cells (HS-5), the membrane anchored ALP catalyzes the formation of the nanofibers on the surface of HeLa cells, but the ACP in the lysosomes of HS-5 generates fluorescent nanofibers mainly inside lysosomes () [Citation54]. Recently, Wang et al. developed an enzyme substrate (17) that targeting lysosome of cells in a more precise fashion [Citation41]. 17, containing a protected tyrosine phosphate, can only be deprotected in acidic environment to expose the phosphate group for further dephosphorylation by ACP, generating 18 (). Such a design prevents undesired dephosphorylation at the cell membrane and cytoplasm, allowing only ACP catalyzed dephosphorylation to occur in lysosomes (). Considering other hydrolytic enzymes inside lysosome, this work implies that it should be feasible to design a substrate that responds to multiple enzymes for targeting lysosome of cells.
ER, as the largest cellular organelle, carry out essential cellular functions, such as protein synthesis, sensing, and post-translational modifications [Citation55]. Several small molecules, such as tunicamycin and thapsigargin, can target ER but lack cell selectivity [Citation56]. Although endogenous proteins rely on a tetrapeptide motif, KDEL, for ER retention [Citation57], conjugating KDEL to other molecules for targeting ER from outside cells is problematic due to the proteolytic susceptibility of the short peptides. It turns out that EISA is quite effective for targeting ER of cancer cells. Specifically, a phosphotetrapeptide (19), containing self-assembling backbone, D-phosphotyrosine, and an L-homoarginine, can disrupt cell membrane and target the ER, resulting in cancer cell death () [Citation42]. The dephosphorylation of 19 generates a tetrapeptide derivative (20). 20 self-assembles into crescent-shaped aggregates on the cancer cell surface (), disrupting the integrity of the cells to allow more 19 enter cells and localizes at ER. A later work [Citation58] suggests that PTPN1 on the outer membrane of ER likely contribute to turn 19 to 20 on ER for enhanced ER targeting.
Although organelle-targeted therapies focused on the highly sensitive and precise attack on specific organelles have made considerable progress for targeting nucleus, mitochondria, lysosome, and ER, the selective targeting of Golgi remains elusive. Two recent studies have demonstrated that enzymatic formation of nanostructures at Golgi is effective for Golgi-targeting. In the first case, using a sulfur atom to replace the oxygen atom in the phosphoester bond of a phosphopeptide produce a thiophosphate peptide (21) () [Citation44]. As expected, ALP dephosphorylated 21 rapidly to form the nanofibers made of 22 (). But unexpectedly, 22 instantly accumulates at Golgi () of the HeLa cells at the concentration as low as 500 nM. Further examination using multiple cell lines reveals ALP-catalyzed Golgi-targeting is cell selective. The fast accumulation of 22 at Golgi occurs in the cells have a high expression level of ALP. Even more unexpectedly, a thioester containing peptide (23), obtained by replacing thiophosphotyrosine with a thioester (), can target Golgi of metazoan cells () [Citation45]. 23 enters cells via caveolin-mediated endocytosis or macropinocytosis and undergoes hydrolysis catalysed by Golgi-associated thioesterases to form the thiopeptide (22) (), which accumulates in the Golgi of metazoan cells unselectively. These results illustrate that using enzyme to generate nanostructure at the Golgi represents an effective and versatile strategy for targeting Golgi according to the need of applications.
Extracellular nanostructures
Unlike most of the membrane-anchored enzymes that orient the active sites at the cytosolic side of the membrane, the catalytic domain of ALP is on the extracellular side of the membrane. That is, ALP can act as an ectoenzyme, catalyzing the formation nanostructures outside cells. This notion turns out to be valid, as demonstrated by several representative examples. In a study examining the response of HeLa cells to a D-phosphotripeptide derivative (24), consisting of a naphthyl capped tripeptide, D-Phe-D-Phe-D-Tyr, ALP can catalytically dephosphorylates 24 to form a hydrogelator (25) () [Citation59]. This hydrogelator self-assembles, resulting in a network of nanofibrils as the scaffold for a hydrogel. Unexpectedly, hydrogelation occurred on the HeLa surface when incubating HeLa cells with 25, as well as in the culture medium (). However, incubating 25 with HeLa cell fails to produce the phenomena observed with 24 (). Scanning electron microscopy (SEM) reveals nanofibers present on the surface of HeLa cells incubated with 24 but not on untreated HeLa cells (). Transmission electron microscope (TEM) image of the pericellular hydrogel on HeLa cells treated by 24 further confirm networks of nanofibers (). Moreover, 24 hardly inhibits HS-5 cells, a cell lacking surface ALP. These results demonstrates that nanofibers of 25, formed by ectoenzyme-instructed self-assembly, selectively inhibit cancer cells. Enzymatic dephosphorylation enables pericellular nanofibers and subsequent hydrogelation. This concept is not only applicable to phosphopeptides, but also works on the phosphosaccharides, as reported by Ulijn et al. [Citation60]
Figure 5. (a) Chemical structures of 24 and 25 and the enzyme-catalyzed formation of pericellular hydrogel to induce cell death. (b) Optical images of the HeLa cells incubated with 24 (560, 280 and 140 μM) or with 25 (560 μM) for 2 h. White arrows point at the hydrogelation site. (c) SEM images of freeze-dried HeLa cells treated with 24 (560 μM) for 2 h. Scale bars = 10 μm. (d) Negative stained TEM images of the pericellular hydrogels on the HeLa cells treated by 24 (280 μM) and the high magnification image of the nanofibrils. Scale bars = 100 nm. (e) CLSM images of different cell lines incubated with 15. The incubation time was 24 h for HS-5, PC3, U-87 MG, Capan-2, A375, SKOV3, PC-12 Adh, and T98G and 12 h for Saos-2 and 7F2. The incubation concentration of 15 was 100 μM for Saos-2 and 7F2 and 500 μM for other cells. Scale bars = 10 μm. (f) CLSM images of HeLa cells incubated with 15 (500 μM, 30 min, 1 h, 3 h and 6 h). Scale bar = 5 μm. (g) CLSM images of two pairs of drug-sensitive and drug-resistant cancer cell lines (A2780 and A2780cis; MES-SA and MES-SA/Dx5, 24 h) incubated with 15, and CLSM images of MCF-7 incubated with 15 without (upper) and with (bottom) the addition of prednisolone. Scale bar = 5 μm. (h) Merged image (differential interference contrast and fluorescence microscopy images) of treated HeLa cells (grown on marked Aclar discs and incubated for 6 h with 500 μM of 15), recorded only a few minutes before the sample was high-pressure frozen for TEM and the high-magnification electron microscopy image shows a slice through the same cell. Scale bar = 2 μm. (i) Higher-magnification electron micrograph of the cell region highlighted by the red box in (H) and the high-magnification electron micrographs of the red boxed area. Scale bar = 200 nm.
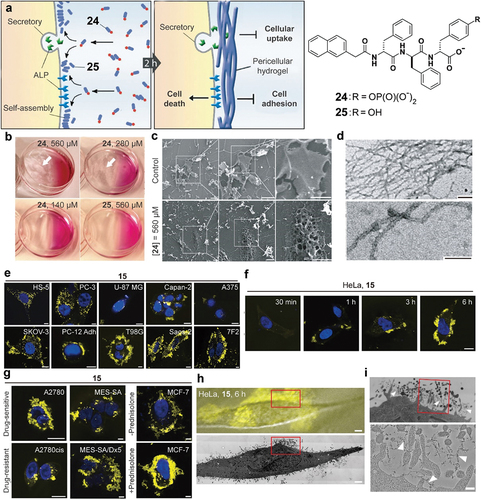
Because the nanofibers of 16 are non-diffusive, they localize on the cell surface, accumulate with time, and exhibit fluorescence with increasing intensity. Thus, the enzymatic conversion of 15 to 16 can be used to assess the activity of ALP on different cell surfaces () [Citation54]. In fact, mammalian ALP, as membrane glycosylphosphatidylinositol (GPI)-anchored ectoenzymes [Citation61], play a critical role from embryogenesis to cancer biology [Citation62]. The spatiotemporal determination of the activities of ALPs of live cells would provide details of dephosphorylation kinetics in cellular processes. Moreover, 16, as a D-peptide, form proteolytic resistant nanofibrils around cancer cells, conferring excellent spatiotemporal resolution to reveal the activity of ALP on cancer cell surface () [Citation54]. As the substrate for EISA, 15 reveals the difference of ALP activities between two pairs of drug sensitive/resistant cancer cell lines (e.g. A2780 and A2780cis, MES-SA and MES-SA/dx5) and a cell line (e.g. MCF-7) under hormonal stimulus (). In addition, correlative light and transmission electron microscopy (CLEM) imaging of the nanofibrils on cells has confirmed the formation of pericellular nanofibers of 16 on HeLa cells (). These results also provide insights to understand the inhibition of HeLa cells by pericellular nanofibers [Citation63].
Intercellular nanostructures
In various of tissues or organs, extracellular matrix (ECM) localizes at intercellular space to interact with cells in a three-dimensional manner. While it is well established that a series of enzymatic reactions leads to the formation of ECM, the use of enzymatic reaction to generate synthetic nanostructures intercellularly is just at its beginning. In a study that aimed to mimic the dynamic post-translation features of ECM, such as glycosylation and phosphorylation, the authors tried to sidestep the difficulty of synthesizing glycoproteins and phosphoproteins by generating supramolecular phosphoglycopeptides that contain a phosphopeptide (26) and a glycopeptide (27) () [Citation64]. The mixture of 26 and 27 self-assembles to form oligomers, which exist as nanoparticles. Adding ALP to the solution of the mixture turns the nanoparticles into nanofibrils (), albeit that the dephosphorylation of 26 to form 28 is incomplete. Circular dichroism spectra suggest that the assemblies of 26 and 27 are dynamic (i.e. reversible binding between 26 (or 28) with 27) and reactive (i.e. 26 as substrates of ALP). When the mixture of 26 and 27 is incubated with the mixture of Saos-2 cells and HS-5 cells [Citation65], a fibroblast cell line, the co-cultured cells spontaneously transform from a 2D cell sheet to 3D spheroids (). Using the medium without 26 and 27 is able to revert the cell spheroids back to the 2D cell sheet. These results indicate that the intercellular assemblies, formed by dephosphorylation, maintain the 3D spheroids.
Figure 6. (a) Chemical structures of 26 and 27 and 28. (b) TEM images of 26:27 (300 μM) after treatment with ALP (0.5 or 0.2 U/mL) for 24 h. Scale bar = 50 nm. (c) Formation of 3D cell spheroids composed of HS-5 and Saos-2 cells from a 2D cell sheet upon the addition of 26:27 (300 μM) for 48 h. Scale bar = 150 μm. (d) Chemical structures of 29 and 30. (e) TEM images of 29 (0.1 wt %) without or with the treatment of ALP (1 U/mL). Scale bar = 50 nm. (f) Optical images of HS-5 cells in culture medium and coincubation with 29 (200 μM) for 24 h. Scale bar = 150 μm. (g) Chemical structures of 31, 32, 33 and 34. (h) 3D reconstruction and cross section and side view of 33 class 1 filaments, 33 class 2 filaments and 34 filaments. (i) Representative confocal images of the FRAP assay on suspended HS-5 cells incubated with 31 (200 µM) for 48 h and quantitative analysis of the fluorescence intensity of the FRAP assay. (j) Confocal images of suspended HS-5 cells with 31 (200 µM) addition under different treatments for 48 h: Control (31 only); + verapamil (Ver) (31 + 20 µM Ver); + brefeldin a (BFA) (31 + 50 nM BFA); + vacuolin-1 (V-1) (31 + 10 µM V-1). Scale bar = 100 μm. (k) TEM images of 32 (50 µM) treated with ALP (0.1 U/mL) for 48 h and then incubated with 50 µg/mL of fibronectin.
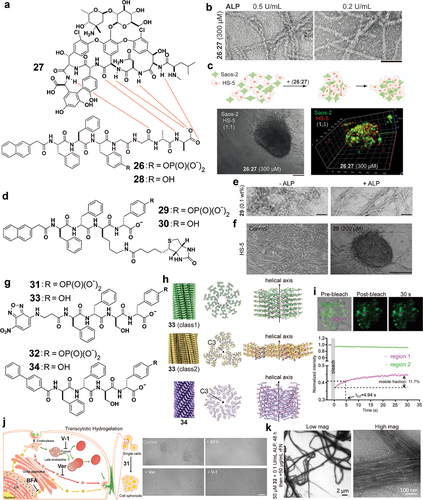
Although 27 is necessary for the mixture of 26 and 27 to enable HS-5 transform from 2D cell culture to 3D cell spheroids, the role of 27 likely is to binding ECM between cells because the conjugation of biotin to a D-phosphopeptide can also induce cell spheroids of HS-5 cells () [Citation66]. In that study, biotin acts as the proteolytic-resistant cell adhesion motif because various cell lines express a high level of biotin receptor [Citation67]. Specifically, a biotinylated D-phosphotetrapeptide (29) serves as the substrate for phosphatase-instructed self-assembly. Being partially dephosphorylated by phosphatases at intercellular space, nanoparticles of 29 turns into nanofibers of the mixture of 29 and 30 at the intercellular space (). This dynamic transformation of the assemblies of 29 induces 3D cell spheroids of HS-5 from a 2D cell sheet (). In this study of 29 for inducing cell spheroids, dephosphorylation, cell surface-binding motif, and proteolytic resistance of the precursor, are essential for the observed cell spheroids.
Although the above two studies have shown that enzyme-responsive D- phosphopeptide assemblies induce HS-5 cell spheroids, the molecular mechanism remained obscure until a recent study, which reveals the transcytosis of the enzyme-responsive D-phosphopeptides forming intercellular D-peptide nanofibers to enhance fibrillogenesis of fibronectin for cell spheroid formation. In that relatively comprehensive study, two D-phosphopeptides were examined. One is NBD-ffspy (31), the other is BP-ffspy (32). Dephosphorylation of 31 or 32 generates the nanofibers of NBD-ffsy (33) or BP-ffsy (34), respectively () [Citation68]. Cryo-EM imaging and reference-free 2D classifications of 33 confirms polymorphic cross-β filaments of 33 with four distinct species. The most dominant filament species has a helical rise of 0.48 Å and twist of 35.8°, with resolution of ~3.1 Å. Another class filaments reached ~3.2 Å resolution with a helical rise of 4.80 Å and a twist of −1.7°. The filament model possesses C3 symmetry, and each asymmetrical unit contains six copies of 33 molecules. The dephosphorylation of 32 forms homogenous filaments of 34 that reach ~2.6 Å resolution with the hydrophobic biphenyl motifs arranging at the center with C3 symmetry, a helical rise of 2.11 Å and twist of −54.8° (). Fluorescence recovery after photobleaching (FRAP) indicates intercellular hydrogelation (). In addition, endocytosis inhibitors or exocytosis inhibitors of distinct pathways significantly reduce cellular uptake of 31 and decrease sizes of spheroids, suggesting the crucial role of transcytosis in spheroid formation (). Moreover, at cell-free condition, incubating rhodamine-labelled fibronectin (rFN) with nanofibers of 33 or 34, facilitates the fibrillogenesis of rFN. TEM imaging also reveals that 31 or 32 with rFN in the presence of ALP results in wider nanofibers with helical features (). According to cryo-EM structures, the filaments of 33 or 34 display C-terminal carboxylic acid and hydroxyl groups on the surface, which are similar to the surface of the filament of CsgA, which is now to interact with fibronectin [Citation69]. This similarity may account for their ability to interact with fibronectin for inducing fibrillogenesis of fibronectin. These results suggest a transcytosis approach for generation cell spheroids by using enzymatic reactions to generate intercellular nanostructures.
Perspective and outlook
Enzymatic reactions and protein–protein interactions within cells form the fundamental basis for cellular functions, representing a crucial aspect of what defines ‘living’ entities. While extensive research has been dedicated to unraveling the intricacies of molecular interactions among biomacromolecules, less attention has been given to the pivotal role of enzymatic reactions in controlling protein–protein interactions. This oversight is particularly noteworthy when considering the development of synthetic nanoarchitectures within cellular contexts. Within such a context, it is rather striking that a seemingly straightforward process such as dephosphorylation can trigger the targeted generation of nanostructures, exhibiting selectivity towards specific cells and even organelles. The simplicity of a dephosphorylation-driven response serves as a compelling indication that numerous untapped opportunities exist for exploring alternative enzymatic reactions in the creation of intricate nanostructures within the dynamic milieu of cells.
To further explore the potential of enzymatic reaction for creating cellular nanostructures, several limitations remain to be addressed, such as enzyme kinetics [Citation70], dynamics of enzyme locations, and molecular structures of the nanoarchitectures. The dynamic nature of molecular assemblies in vivo presents a challenge in determining enzyme kinetics in cellular environments, especially when multiple enzymes act on the same substrates, or multiple substrates can be transformed by the same enzymes. The recently developed bond-selective imaging of enzymatic reactions [Citation71,Citation72] likely would provide a possible solution to address this challenge. The subcellular locations of enzymes are unlikely to be static but rather are context dependent and dynamic [Citation73,Citation74]. Tracking the enzyme location over a long period of time and a large area in cellular environment is possible for a single enzyme but remains a challenge to measure the location and activity of multiple enzymes simultaneously. To address this issue, super resolution fluorescent imaging and electron-tomograph may provide an approach to understand such sophisticated molecular machinery of cells [Citation75,Citation76]. The atomistic structures of functional nanostructures are essential in understanding the structure–activity relationship. The ‘resolution revolution’ of cryo-EM has greatly advanced the structural elucidation of protein and peptide nanofibers, as recently summarized by Egelman et al. [Citation77] The use of cryo-EM for determining the atomistic structures of nanoarchitecture will definitely see increased applications, and many useful insights will be revealed.
Over the last two decades, intracellular peptide nanostructures have received most research attentions because their potential applications in biomedicine. The use of enzymatic reactions to generate exogeneous peptide nanostructures in cells implies the concept should be applicable for generating intracellular nanostructures of other molecular building blocks, such as nucleic acids or carbohydrates, which provide exciting new opportunities. Further applying enzymatic reactions at the intersection of chemistry and cell biology would likely lead to many discoveries. The pathway to future success and groundbreaking discoveries hinges on the precise engineering of molecules to facilitate diverse enzymatic reactions [Citation78–80] within cellular environments. This imperative task demands a harmonious integration of expertise across multiple disciplines, bringing together the skills and insights of chemists, biologists, engineers, and medicinal scientists. By fostering collaborative efforts among these diverse fields, we will unlock the potential to unravel the complexities of cellular processes and pave the way for transformative advancements in science and technology.
Statement of novelty
Enzymatic reactions in cells create precise nanoarchitectures, offering insights into cell biology through controllable nanoarchitectonics, as shown by numerous examples in this review.
Acknowledgments
This work is partially supported by NIH CA142746 & CA262920.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
Notes on contributors

Weiyi Tan
Weiyi Tan obtained his BS degree from the College of Chemistry, Chemical Engineering and Material Science, Soochow University, in 2018. He is currently in his sixth year as a graduate student in chemistry supervised by Professor Bing Xu at Brandeis University. His current research interest lies in designing self-assembling materials for biological applications.

Qiuxin Zhang
Qiuxin Zhang obtained her BEng degree in Nanomaterials and Nanotechnology from the College of Nano Science and Technology, Soochow University and BS degree in Materials and Nanosciences from University of Waterloo both in 2019. She is currently a fifth-year graduate student supervised by Prof. Bing Xu in the Department of Chemistry at Brandeis University. Her research interest focuses on peptide assemblies and their promise in cancer therapeutics.

Mikki Lee
Mikki Lee is a third-year undergraduate student at the National University of Singapore, where she is pursuing a double major in Pharmaceutical Science and Life Science. Mikki’s research interest lies in advancing drug delivery technologies and nanotherapeutics for cancer therapy.

William Lau
William Lau is a rising senior majoring in chemical biology. William is currently interested in biotech research, especially in the field of organic chemistry in oncology research.

Bing Xu
Prof. Bing Xu after receiving his BS and MS from Nanjing University in 1987 and 1990, respectively, obtained his PhD in 1996 from the University of Pennsylvania. Before starting his independent research at the Hong Kong University of Science and Technology from 2000, he was an NIH postdoctoral fellow at Harvard University. He was tenured as an associate professor in January 2006 and became a full professor in July 2008 at HKUST. He is currently a professor at the Department of Chemistry, Brandeis University. His current research focuses on the applications of enzymatic noncovalent synthesis in materials, biology, and medicine.
References
- Whitesides GM, Mathias JP, Seto CT. Molecular self-assembly and nanochemistry: a chemical strategy for the synthesis of nanostructures. Sci. 1991;254(5036):1312–15. doi: 10.1126/science.1962191
- He H, Tan W, Guo J, et al. Enzymatic noncovalent synthesis. Chem Rev. 2020 2020/09/23;120(18):9994–10078. doi: 10.1021/acs.chemrev.0c00306
- Iwasaki Y, Yamane T. Enzymatic synthesis of structured lipids. Recent progress of biochemical and biomedical engineering in Japan I. Berlin, Heidelberg: Springer Berlin Heidelberg; 2004. p. 151–171.
- Coumans JVF, Davey RJ, Moens PDJ. Cofilin and profilin: partners in cancer aggressiveness. Biophys Rev. 2018 2018/10/01;10(5):1323–1335. doi: 10.1007/s12551-018-0445-0
- Almanza A, Carlesso A, Chintha C, et al. Endoplasmic reticulum stress signalling – from basic mechanisms to clinical applications. FEBS J. 2019;286(2):241–278. doi: 10.1111/febs.14608
- Lu A, Magupalli Venkat V, Ruan J, et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014;156(6):1193–1206. doi: 10.1016/j.cell.2014.02.008
- Zhang Y, Luo L, Xu X, et al. Acetylation is required for full activation of the NLRP3 inflammasome. Nat Commun. 2023 2023/12/18;14(1):8396. doi: 10.1038/s41467-023-44203-0
- Zheng S, Que X, Wang S, et al. ZDHHC5-mediated NLRP3 palmitoylation promotes NLRP3-NEK7 interaction and inflammasome activation. Molecular Cell. 2023;83(24):4570–4585.e7. doi: 10.1016/j.molcel.2023.11.015
- Yang Z, Liang G, Xu B. Enzymatic hydrogelation of small molecules. Acc Chem Res. 2008 Feb;41(2):315–326. doi: 10.1021/ar7001914
- Gao Y, Yang Z, Kuang Y, et al. Enzyme-instructed self-assembly of peptide derivatives to form nanofibers and hydrogels. Biopolym. 2010;94(1):19–31. doi: 10.1002/bip.21321
- Zhou J, Xu B. Enzyme-instructed self-assembly: a multistep process for potential cancer therapy. Bioconjug Chem. 2015 Jun 17;26(6):987–999. doi: 10.1021/acs.bioconjchem.5b00196
- Nagarajan R. Molecular packing parameter and surfactant self-assembly: the neglected role of the surfactant tail. Langmuir: The ACS J Surfaces And Colloids. 2002 2002/01/01;18(1):31–38.
- Yang Z, Gu H, Fu D, et al. Enzymatic formation of supramolecular hydrogels. Adv Mater. 2004;16(16):1440–1444. doi: 10.1002/adma.200400340
- Toledano S, Williams RJ, Jayawarna V, et al. Enzyme-triggered self-assembly of peptide hydrogels via reversed hydrolysis. J Am Chem Soc. 2006 2006/02/01;128(4):1070–1071. doi: 10.1021/ja056549l
- Tirado P, Reisch A, Roger E, et al. Catalytic saloplastics: alkaline phosphatase immobilized and stabilized in compacted polyelectrolyte complexes. Adv Funct Materials. 2013;23(38):4785–4792. doi: 10.1002/adfm.201300117
- Vigier-Carrière C, Garnier T, Wagner D, et al. Bioactive seed layer for surface-confined self-assembly of peptides. Angewandte Chemie. 2015;54(35):10198–10201. doi: 10.1002/anie.201504761
- Rodon Fores J, Criado-Gonzalez M, Chaumont A, et al. Autonomous growth of a spatially localized supramolecular hydrogel with autocatalytic ability. Angewandte Chemie. 2020;59(34):14558–14563. doi: 10.1002/anie.202005377
- Muller C, Ontani A, Bigo-Simon A, et al. Localized enzyme-assisted self-assembly of low molecular weight hydrogelators. Mechanism, applications and perspectives. Adv Colloid Interface Sci. 2022;304:102660. doi: 10.1016/j.cis.2022.102660
- Runser J-Y, Fneich F, Senger B, et al. Transition from continuous to microglobular shaped peptide assemblies through a Liesegang-like enzyme-assisted mechanism. J Colloid Interface Sci. 2023;633:876–885. doi: 10.1016/j.jcis.2022.11.034
- Yang Z, Liang G, Wang L, et al. Using a kinase/phosphatase switch to regulate a supramolecular hydrogel and forming the supramolecular hydrogel in vivo. J Am Chem Soc. 2006 Mar 8;128(9):3038–3043. doi: 10.1021/ja057412y
- Yang Z, Liang G, Guo Z, et al. Intracellular hydrogelation of small molecules inhibits bacterial growth. Angew Chem Int Ed. 2007;46(43):8216–8219. doi: 10.1002/anie.200701697
- Reches M, Gazit E. Casting metal nanowires within discrete self-assembled peptide nanotubes. Science. 2003;300(5619):625–627. doi: 10.1126/science.1082387
- Mahler A, Reches M, Rechter M, et al. Rigid, self-assembled hydrogel composed of a modified aromatic dipeptide. Advan Mater. 2006 Jun 6;18(11):1365–1370. doi: 10.1002/adma.200501765
- Yang Z, Liang G, Guo Z, et al. Intracellular hydrogelation of small molecules inhibits bacterial growth. Angewandte Chemie. 2007;46(43):8216–8219. doi: 10.1002/anie.200701697
- Yang ZM, Xu KM, Guo ZF, et al. Intracellular enzymatic formation of nanofibers results in hydrogelation and regulated cell death. Advan Mater. 2007 Oct 19;19(20):3152–3156. doi: 10.1002/adma.200701971
- Liang G, Ren H, Rao J. A biocompatible condensation reaction for controlled assembly of nanostructures in living cells. Nat Chem. 2010 2010/01/01;2(1):54–60. doi: 10.1038/nchem.480
- Zheng D, Liu J, Ding Y, et al. Tandem molecular self-assembly for selective lung cancer therapy with an increase in efficiency by two orders of magnitude. Nanoscale. 2021;13(24):10891–10897. doi: 10.1039/D1NR01174J
- Yang J, An H-W, Wang H. Self-assembled peptide drug delivery systems. ACS Appl Bio Mater. 2021 2021/01/18;4(1):24–46. doi: 10.1021/acsabm.0c00707
- Hu Y, Zhang J, Miao Y, et al. Enzyme-mediated in situ self-assembly promotes in vivo bioorthogonal reaction for pretargeted multimodality imaging. Angewandte Chemie. 2021;60(33):18082–18093. doi: 10.1002/anie.202103307
- Feng Z, Wang H, Wang F, et al. Artificial intracellular filaments. Cell Rep Phys Sci. 2020 Jul 22;1(7). doi: 10.1016/j.xcrp.2020.100085
- Egelman EH. Ambiguities in helical reconstruction [Article]. Elife. 2014 Dec;3:e04969. doi: 10.7554/eLife.04969
- Feng Z, Han X, Wang H, et al. Enzyme-instructed peptide assemblies selectively inhibit bone tumors. Chem. 2019 Sep 12;5(9):2442–2449. doi: 10.1016/j.chempr.2019.06.020
- Zhang Y, Kuang Y, Gao Y, et al. Versatile small-molecule motifs for self-assembly in water and the formation of biofunctional supramolecular hydrogels. Langmuir. 2011 Jan 18;27(2):529–537. doi: 10.1021/la1020324
- Wang H, Feng Z, Wang Y, et al. Integrating enzymatic self-assembly and mitochondria targeting for selectively killing cancer cells without acquired drug resistance. J Am Chem Soc. 2016 2016/12/14;138(49):16046–16055. doi: 10.1021/jacs.6b09783
- He H, Wang J, Wang H, et al. Enzymatic cleavage of branched peptides for targeting mitochondria. J Am Chem Soc. 2018 2018/01/31;140(4):1215–1218. doi: 10.1021/jacs.7b11582
- Yang L, Peltier R, Zhang M, et al. Desuccinylation-triggered peptide self-assembly: live cell imaging of SIRT5 activity and mitochondrial activity modulation. J Am Chem Soc. 2020 2020/10/21;142(42):18150–18159. doi: 10.1021/jacs.0c08463
- Jeena MT, Palanikumar L, Go EM, et al. Mitochondria localization induced self-assembly of peptide amphiphiles for cellular dysfunction. Nat Commun. 2017 2017/06/21;8(1):26. doi: 10.1038/s41467-017-00047-z
- Ji S, Li J, Duan X, et al. Targeted enrichment of enzyme-instructed assemblies in cancer cell lysosomes turns immunologically cold tumors hot. Angewandte Chemie. 2021;60(52):26994–27004. doi: 10.1002/anie.202110512
- Wang J, Hu L, Zhang H, et al. Intracellular condensates of oligopeptide for targeting lysosome and addressing multiple drug resistance of cancer. Adv Mater. 2022;34(1):2104704. doi: 10.1002/adma.202104704
- Hu L, Li Y, Lin X, et al. Structure-based programming of supramolecular assemblies in living cells for selective cancer cell inhibition. Angewandte Chemie. 2021;60(40):21807–21816. doi: 10.1002/anie.202103507
- Yang X, Lu H, Tao Y, et al. Spatiotemporal control over chemical assembly in living cells by integration of acid-catalyzed hydrolysis and enzymatic reactions. Angewandte Chemie. 2021;60(44):23797–23804. doi: 10.1002/anie.202109729
- Feng Z, Wang H, Wang S, et al. Enzymatic assemblies disrupt the membrane and target endoplasmic reticulum for selective cancer cell death. J Am Chem Soc. 2018 2018/08/01;140(30):9566–9573. doi: 10.1021/jacs.8b04641
- Gan S, Yang L, Heng Y, et al. Enzyme-directed and organelle-specific sphere-to-fiber nanotransformation enhances photodynamic therapy in cancer cells. Small Met. 2024:2301551. doi: 10.1002/smtd.202301551
- Tan W, Zhang Q, Wang J, et al. Enzymatic assemblies of thiophosphopeptides instantly target golgi apparatus and selectively kill cancer cells**. Angewandte Chemie. 2021;60(23):12796–12801. doi: 10.1002/anie.202102601
- Tan W, Zhang Q, Quiñones- FríFríAs MC, et al. Enzyme-responsive peptide thioesters for targeting golgi apparatus. J Am Chem Soc. 2022 2022/04/20;144(15):6709–6713. doi: 10.1021/jacs.2c02238
- Lipsky NG, Pagano RE. A vital stain for the golgi apparatus. Sci. 1985;228(4700):745–747. doi: 10.1126/science.2581316
- Ishida M, Watanabe H, Takigawa K, et al. Synthetic self-localizing ligands that control the spatial location of proteins in living cells. J Am Chem Soc. 2013 2013/08/28;135(34):12684–12689. doi: 10.1021/ja4046907
- Zhu H, Oh JH, Matsuda Y, et al. Tyrosinase-based proximity labeling in living cells and in vivo. J Am Chem Soc. 2024 2024/03/20;146(11):7515–7523. doi: 10.1021/jacs.3c13183
- Smith RAJ, Porteous CM, Gane AM, et al. Delivery of bioactive molecules to mitochondria in vivo. Proc Natl Acad Sci. 2003;100(9):5407–5412.
- Prag HA, Kula-Alwar D, Pala L, et al. Selective delivery of dicarboxylates to mitochondria by conjugation to a lipophilic cation via a cleavable linker. Mol Pharm. 2020 2020/09/08;17(9):3526–3540. doi: 10.1021/acs.molpharmaceut.0c00533
- Hopp TP, Prickett KS, Price VL, et al. A short polypeptide marker sequence useful for recombinant protein identification and purification. Nat Biotechnol. 1988;6(10):1204–1210. doi: 10.1038/nbt1088-1204
- He H, Lin X, Wu D, et al. Enzymatic noncovalent synthesis for mitochondrial genetic engineering of cancer cells. Cell Rep Phys Sci. 2020 2020/12/23/;1(12):100270. doi: 10.1016/j.xcrp.2020.100270
- Saftig P, Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol. 2009 2009/09/01;10(9):623–635. doi: 10.1038/nrm2745
- Zhou J, Du X, Berciu C, et al. Enzyme-instructed self-assembly for spatiotemporal profiling of the activities of alkaline phosphatases on live cells. Chem. 2016 2016/08/11/;1(2):246–263. doi: 10.1016/j.chempr.2016.07.003
- Voeltz GK, Rolls MM, Rapoport TA. Structural organization of the endoplasmic reticulum. EMBO Rep. 2002;3(10):944–950. doi: 10.1093/embo-reports/kvf202
- Zhang X, Yuan Y, Jiang L, et al. Endoplasmic reticulum stress induced by tunicamycin and thapsigargin protects against transient ischemic brain injury. Autophagy. 2014 2014/10/17;10(10):1801–1813. doi: 10.4161/auto.32136
- Stornaiuolo M, Lotti LV, Borgese N, et al. KDEL and KKXX retrieval signals appended to the same reporter protein determine different trafficking between endoplasmic reticulum, intermediate compartment, and golgi complex. Mol Biol Cell. 2003;14(3):889–902. doi: 10.1091/mbc.e02-08-0468
- Feng Z, Wang H, Xu B. Instructed assembly of peptides for intracellular enzyme sequestration. J Am Chem Soc. 2018 Dec 5;140(48):16433–16437. doi: 10.1021/jacs.8b10542
- Kuang Y, Shi J, Li J, et al. Pericellular hydrogel/nanonets inhibit cancer cells. Angewandte Chemie. 2014;53(31):8104–8107. doi: 10.1002/anie.201402216
- Pires RA, Abul-Haija YM, Costa DS, et al. Controlling cancer cell fate using localized biocatalytic self-assembly of an aromatic carbohydrate amphiphile. J Am Chem Soc. 2015 2015/01/21;137(2):576–579. doi: 10.1021/ja5111893
- Sesana S, Re F, Bulbarelli A, et al. Membrane features and activity of GPI-anchored enzymes: alkaline phosphatase reconstituted in model membranes. Biochem. 2008 2008/05/01;47(19):5433–5440. doi: 10.1021/bi800005s
- Yalak G, Ehrlich YH, Olsen BR. Ecto-protein kinases and phosphatases: an emerging field for translational medicine. J Transl Med. 2014 2014/06/12;12(1):165. doi: 10.1186/1479-5876-12-165
- Du X, Zhou J, Wang H, et al. In situ generated D-peptidic nanofibrils as multifaceted apoptotic inducers to target cancer cells. Cell Death Dis. 2017 Feb 16;8(2):e2614. doi: 10.1038/cddis.2016.466
- Wang H, Feng Z, Xu B. Instructed assembly as context-dependent signaling for the death and morphogenesis of cells. Angewandte Chemie. 2019;58(17):5567–5571. doi: 10.1002/anie.201812998
- Roecklein BA, Torokstorb B. Functionally distinct human marrow stromal cell lines immortalized by transduction with the human papilloma virus E6/E7 genes. Blood. 1995 Feb;85(4):997–1005. doi: 10.1182/blood.V85.4.997.bloodjournal854997
- Wang H, Feng Z, Xu B. Intercellular instructed-assembly mimics protein dynamics to induce cell spheroids. J Am Chem Soc. 2019 May 8;141(18):7271–7274. doi: 10.1021/jacs.9b03346
- Russell-Jones G, McTavish K, McEwan J, et al. Vitamin-mediated targeting as a potential mechanism to increase drug uptake by tumours. J Inorg Biochem. 2004;98(10):1625–1633. doi: 10.1016/j.jinorgbio.2004.07.009
- Guo J, Wang F, Huang Y, et al. Cell spheroid creation by transcytotic intercellular gelation. Nat Nanotechnol. 2023 2023/09/01;18(9):1094–1104. doi: 10.1038/s41565-023-01401-7
- Loferer H, Hammar M, Normark S. Availability of the fibre subunit CsgA and the nucleator protein CsgB during assembly of fibronectin-binding curli is limited by the intracellular concentration of the novel lipoprotein CsgG. Mol Microbiol. 1997;26(1):11–23. doi: 10.1046/j.1365-2958.1997.5231883.x
- Qiao Y, Wu G, Liu Z, et al. Assessment of the enzymatic dephosphorylation kinetics in the assemblies of a phosphopentapeptide that forms intranuclear nanoribbons. Biomacromolecul. 2024 2024/02/12;25(2):1310–1318. doi: 10.1021/acs.biomac.3c01288
- He H, Yin J, Li M, et al. Mapping enzyme activity in living systems by real-time mid-infrared photothermal imaging of nitrile chameleons. Nat Methods. 2024 2024/02/01;21(2):342–352.
- Zhao J, Matlock A, Zhu H, et al. Bond-selective intensity diffraction tomography. Nat Commun. 2022 2022/12/15;13(1):7767. doi: 10.1038/s41467-022-35329-8
- Yi M, Wang F, Tan W, et al. Enzyme responsive rigid-rod aromatics target “undruggable” phosphatases to kill cancer cells in a mimetic bone microenvironment. J Am Chem Soc. 2022 2022/07/27;144(29):13055–13059. doi: 10.1021/jacs.2c05491
- Yi M, Ashton-Rickardt G, Tan W, et al. Accelerating cellular uptake with unnatural amino acid for inhibiting immunosuppressive cancer cells. Chem – A Eur J. 2024;30:e202400691.
- Zhang C, Tian Z, Chen R, et al. Advanced imaging techniques for tracking drug dynamics at the subcellular level. Adv Drug Deliv Rev. 2023 2023/08/01/;199:114978. doi: 10.1016/j.addr.2023.114978
- Baumeister W. Cryo-electron tomography: the power of seeing the whole picture. Biochem Biophys Res Commun. 2022 2022/12/10/;633:26–28. doi: 10.1016/j.bbrc.2022.08.078
- Wang F, Gnewou O, Solemanifar A, et al. Cryo-EM of helical polymers [review]. Chem Rev. 2022;122(17):14055–14065. doi: 10.1021/acs.chemrev.1c00753
- Chen N, Zhang Z, Liu X, et al. Sulfatase-induced in situ formulation of antineoplastic supra-PROTACs. J Am Chem Soc. 2024 2024/04/17;146(15):10753–10766. doi: 10.1021/jacs.4c00826
- Tanaka A, Fukuoka Y, Morimoto Y, et al. Cancer cell death induced by the intracellular self-assembly of an enzyme-responsive supramolecular gelator. J Am Chem Soc. 2015 2015/01/21;137(2):770–775. doi: 10.1021/ja510156v
- Morita K, Nishimura K, Yamamoto S, et al. In situ synthesis of an anticancer peptide amphiphile using tyrosine kinase overexpressed in cancer cells. JACS Au. 2022 2022/09/26;2(9):2023–2028. doi: 10.1021/jacsau.2c00301