1. Driver mutations, cancer, and detection methods
Mutation is the permanent change in the DNA sequence of a genome. Mutations can alter the function of encoded proteins with pathophysiological consequences. Driver mutations are a type of gene alteration that can cause dysregulated cell division and give a growth advantage, ultimately leading to tumor development and progression [Citation1,Citation2]. Driver mutations are a hallmark of cancer. For example, mutations in BRCA1/2 genes are closely correlated with breast cancer risk [Citation3]; lung cancers bearing mutated EGFR respond to EGFR inhibitors [Citation4,Citation5]; and melanomas bearing mutated BRAF usually respond to BRAF inhibitors [Citation6,Citation7]. As driver mutations are cancer biomarkers, they have been targeted in precision oncology, for cancer diagnosis, tailored therapy, and treatment response monitoring [Citation8,Citation9].
Molecular cancer diagnostics requires sensitive detection of driver mutations at a very low frequency in early stages of cancer, and accurate quantification of mutation frequency in a complex biological sample [Citation10]. Although many methods have been developed for the detection of mutation [Citation11,Citation12], there are still limitations in terms of clinically availability, sensitivity, specificity, and efficiency. Currently, PCR-based amplification methods including allele-specific PCR and digital PCR are widely used to detect cancer gene mutation in the clinic setting. These technologies require expensive instrumentation, reagents, well-trained personnel and standardized laboratory protocols, and the results may be compromised by contamination [Citation13,Citation14]. The next generation sequencing (NGS) technology is a most promising method for high throughput cancer genome profiling and residual disease detection. However, its inherent weakness such as long turn-around-time (usually several weeks), high cost, requirement of adequate tissue source of nucleic acids, sequencing library preparation, complex bioinformatics, currently hinders its successful implementation in clinical practice [Citation15–Citation17]. Development of novel strategies for the accurate detection of gene mutations, in particular point mutations (single-base changes) or single nucleotide polymorphism (SNP), is urgently needed [Citation18–Citation20].
In this review, we describe two different single-molecule platforms, nanolock-nanopore [Citation21] ()) and nanocross–nanopore [Citation22,Citation23] biosensers ()) for the detection of point mutations. The test-bed of these studies is the BRAF V600E mutation, which occurs with high prevalence in several types of cancer and is the target of several drugs [Citation24]. Accurate detection of this SNP could help physicians diagnose cancers earlier and treat them with individualized therapies. Using the two nanopore methods, we are able to directly ‘read’ single DNA molecules of the low frequency BRAF V600E allele against a background of the wild-type sequence, and quantify the mutation frequency in tumor tissues of cancer patients.
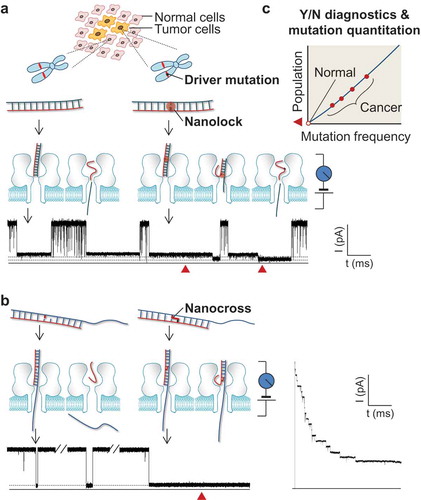
Figure 1. Single-molecule detection of cancer driver mutation by using nanolock-nanapore and nanocross-nanopore sensors. (a) Nanolock-nanopore detection of the BRAF V600E mutant allele in tumor tissue DNA sample; (b) Nanocross-nanopore detection of the BRAF V600E mutant DNA sequence. In both methods, the non-covalent nanolock (a) or covalent nanocross (b) is constructed only on the mutant•probe DNA complex. Their special molecular configuration changes in the nanopore produce a unique ion current signature (current blockade marked by red solid triangle), which serves as a fingerprint of a single mutant DNA molecule. For the nanocross method (b), the unique long-duration blockade by the nanocross enables multi-nanopore simultaneous detection of mutant DNA; (c) Identification of mutant fingerprints enables Yes/No early cancer diagnostics, and counting of mutant fingerprints allows quantitation of mutant frequency for personalized treatment. This figure was adapted with permission from (Wang Y, Tian K, Shi R et al. Nanolock-Nanopore Facilitated Digital Diagnostics of Cancer Driver Mutation in Tumor Tissue. ACS sensors, 2(7), 975–981 (2017)). Copyright (2017) American Chemical Society; adapted with permission from (Zhang X, Price NE, Fang X, Yang Z, Gu LQ, Gates KS. Characterization of Interstrand DNA-DNA Cross-Links Using the alpha-Hemolysin Protein Nanopore. ACS nano, 9(12), 11812–11819 (2015)). Copyright (2015) American Chemical Society; adapted from (Nejad MI, Shi R, Zhang X, Gu LQ, Gates KS. Sequence-Specific Covalent Capture Coupled with High-Contrast Nanopore Detection of a Disease-Derived Nucleic Acid Sequence. ChemBioChem, 18(14), 1383–1386 (2017)) with permission of John Wiley and Sons, Copyright (2017).
2. Nanolock–nanopore and nanocross–nanopore sensors for SNP detections
The sensing element is a nanopore, a 2-nm-wide protein channel that is provided by α-hemolysin embedded in a lipid membrane. A transmembrane voltage is applied to drive an ion current through the pore. When a target molecule in the solution passes through the nanopore, it changes the current in a characteristic manner, thus producing an electric signature that enables single molecule target detection. In the BRAF V600E detection, we used a DNA probe that can hybridize with both mutant and wild-type alleles to form the mutant•probe and wild-type•probe complexes. Powered by the electrophoretic potential of the device, both complexes can be captured by the nanopore and subsequently unzipped by this molecular machine. In order to selectively discriminate the mutant BRAF allele, we have constructed the mutation sequence-specific nanolock and nanocross motifs.
Nanolock is a class of inter-strand stabilizing chemical structures that is formed by binding of a ligand [Citation25–Citation28] at specific base pairs. Fundamentally the nanolock exploits the concept of a stimuli-responsive nucleic acid [Citation29], in which the polymer’s properties are altered in response to an environmental stimulus such as ligand binding. For detection of BRAF V600E [Citation21] ()), we designed a special probe that forms the thymine–thymine (T–T) mismatched base pair in the mutant•probe complex. The T–T mismatch can be bound with a mercuric ion (Hg2+) to form a stabilizing T–Hg2+–T ‘nanolock’ [Citation25,Citation30–Citation33]. In contrast, the wild-type•probe complex does not contain the T–T mismatch and thus cannot form the nanolock. Most importantly, due to the high stability of the nanolock, the mutant•probe unzipping process in the nanopore splits into two consecutive steps that can be resolved in the nanopore. In contrast, the unzipping of the wild-type•probe complex without the nanolock remains a transient, cooperative, one-step process. This mechanistic difference in the unzipping processes allows the locked mutant•probe complex to generate a distinct nanopore signature. The nanolock-produced unzipping signature functions as an ‘electrical fingerprint’, enabling discrimination of the mutant allele from the wild-type sequence. With the fingerprint, we are not only able to ‘read’ each mutant allele molecules, but also quantify the mutation frequency (mutation percentage) in a tumor tissue sample by measuring the occurrence of the nanolock signature.
We have also developed a separate approach that exploits sequence specific cross-linking with a reactive probe strand that covalently captures the mutant BRAF target sequence [Citation22,Citation23] ()). In this ‘nanocross’ approach, a DNA abasic site, embedded in a specialized probe sequence, enables highly selective covalent cross-linking to adenine 1799 in the BRAF V600E mutant gene sequence [Citation34]. Importantly, the cross-linked probe–target complex gives an unmistakable, long-lasting current block in the nanopore device [Citation22,Citation23]. This current signature is easily distinguished from the short-duration blocks caused by uncross-linked target•probe complexes derived from the wild-type sequence. Accordingly, we showed that the fraction of T > A mutant sequence could be quantitatively determined within mixtures containing a high background of the wild-type allele [Citation23].
3 Advantages, improvement, and applications
The nanopore has been broadly investigated for various genetic [Citation35–Citation39], epigenetic [Citation40–Citation43] and proteomic [Citation44–Citation46] detection strategies [Citation47]. Many excellent studies have demonstrated nanopore’s single-nucleotide sensitivity [Citation48–Citation50], and ability to detect single-nucleotide polymorphism [Citation49,Citation51–Citation53]. However, their applications in point mutation detection in clinical setting have not been explored. The nanopore-based next-generation sequencing technology is being developed [Citation52,Citation54–Citation57], but currently the sequencing accuracy (90%) remains impracticable for detecting cancer mutations at low frequencies [Citation58]. By comparison, a great advantage of the nanolock–nanopore and nanocross–nanopore sensors is their high sensitivity. Due to the capability to discriminate each individual mutation DNA molecules captured by the nanopore, the sensor in principle can detect mutant alleles at very low frequency. For BRAF V600E, the sensor has been used to identify mutant alleles at 1% in the presence of wild-type DNA. This sensitivity, which can be further enhanced upon system improvement (see later), is superior to commercially available mutation detection methods such as polymerase chain reaction (PCR) and next generation sequencing (NGS) [Citation21]. In addition to high sensitivity, both nanolock– and nanocross–nanopore sensors exhibit their own advantages. The non-covalent nanolock can be formed immediately upon addition of the ligand in the solution, therefore vastly decreasing the time for sample preparation prior to detection. By comparison, the formation of the covalent nanocross takes much longer time. However, the resulting long-lasting blockade is a unique signature for unmistakable discrimination of the mutant DNA. The nanocross signature can be easily terminated by reversing the voltage polarity (to release the cross-linked DNA duplex into the solution) to capture the next DNA duplex. In addition, due to the nanocross’ long block duration, we can record the time-dependent capture of cross-linked duplexes in a device with multiple nanopores embedded in the membrane. The current trace for such an experiment is characterized by stepwise current drops corresponding to sequential capture of cross-linked duplexes from the test solution (), right current trace). Because the single-molecule capture rate is proportional to the concentration of cross-links in the cis solution and the number of nanopores in the membrane, the multi-channel method will not only vastly shorten the detection time, but also increase the sensitivity, making it useful in particular for detection at low mutant frequency.
To enable future clinical applications, we need improve the performance of both nanopore sensors, including applicability, throughput and sensitivity. First, the nanolock and nanocross toolbox must be expanded in order to target a broader spectrum of driver mutations in human cancers (e.g. mutations on KRAS and EGFR). Among various SNPs, T or A can be detected using the T–Hg2+–T nanolock and nanocross; C or G can be detected using the C–Ag+–C nanolock [Citation25,Citation30–Citation33,Citation43]. In addition, many mismatch-specific chelating compounds [Citation26,Citation27] and metallointercalators [Citation28] with great base-pair selectivity can be utilized to construct mutation-targeted nanolocks. Similarly, additional cross-linking motifs will be developed for the covalent capture of various target sequences. Furthermore, because accurate clinical diagnostics requires simultaneous detection of multiple mutations rather than just one, we will develop methods such as ‘nanopore barcoding’ (exploiting a method we developed for multiple microRNA detection [Citation42]) for simultaneous detection of multiple biomarker mutations. Last, the analytical procedure in the current stage could be slow (low DNA capture rate) due to the low DNA concentrations. To enhance the detection efficiency, one solution is utilizing miniaturized nanopore system such as droplet nanopore [Citation45] and nanopore chips [Citation58]. The miniaturized device allows using very small volume (1 µl) of solution. Therefore the detection can be performed in pre-concentrated and enriched sample with highly elevated single molecule capture rate. Ultimately, it may be possible to realize amplification-free detection in a clinical setting.
Early cancer diagnostics demand detection of mutations at very low frequency. The nanocross– and nanolock–nanopore sensors ultimately may be sufficiently sensitive to execute this task ()). Since this single-molecule detection technology can determine if a DNA molecule is a mutant or wild-type, its false-positive and false-negative rates are both low. Ideally, observation of just one mutant DNA in a given time (e.g. 1 h) would immediately suggests the presence of that specific mutation in the sample. Longer detection time results in higher sensitivity, because the chance to capture a single mutant DNA molecule increases as the detection time increases. Therefore, the techniques, in essence, yield digital signals that are sensitive enough to perform ‘yes/no binary diagnostics’ of a cancer mutation, allowing doctors to quickly take appropriate clinical action.
Recent studies have demonstrated that circulating tumor DNA (ctDNA) fragments carry tumor specific sequence alterations even in the early stages of cancer [Citation59]. Present in the cell-free fraction of blood ‘liquid biopsy’, ctDNA offers a noninvasive ‘real-time’ biomarker of the disease [Citation59–Citation61]. While clinical use of nanolock– and nanocross–nanopore sensors for ctDNA detection remains to be explored, the initial results offer a possibility for a new non-invasive way to assist cancer early diagnostics, potentially leading to improved patient survival and quality of life. The promise of commercially available portable nanopore devices means that this approach could facilitate the early diagnosis of disease in low-resource clinical environments.
Because these nanopore techniques can be used to accurately quantify the frequency of a particular mutation ()), the clinical investigator would be able to use it for monitoring the levels of cancer mutations in patients being treated with particular drugs or other interventions, i.e. monitoring the efficacy of a treatment by measuring changes in the presence of driver mutations before and after administration of a given drug or surgical procedure. This would greatly help doctors in assessing the efficacy of therapy on a patient-by-patient personalized basis.
In summary, the nanolock– and nanocross–nanopore sensors are emerging as a powerful technology with an ability to detect point mutations that occur at very low frequencies. By intuitively observing the unzipping kinetics between nanolock-bound mutated DNA molecules and wild-type molecules in a nanopore or the unmistakable long-duration current blocks associated with cross-links, we are able to differentiate the mutated and wild-type gene sequences. Therefore, this single-molecule-based technology has a potential to be used in cancer diagnostics, personalized medicine, and biomarker discovery.
Declaration of interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties. Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
Additional information
Funding
References
- Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science. 2013;339(6127):1546–1558.
- Lee JS. Exploring cancer genomic data from the cancer genome atlas project. BMB Rep. 2016;49(11):607–611.
- Couch FJ, Nathanson KL, Offit K. Two decades after BRCA: setting paradigms in personalized cancer care and prevention. Science. 2014;343(6178):1466–1470.
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139.
- Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500.
- Bollag G, Hirth P, Tsai J, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467(7315):596–599.
- Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363(9):809–819.
- Bollag G, Tsai J, Zhang J, et al. Vemurafenib: the first drug approved for BRAF-mutant cancer. Nat Rev Drug Discov. 2012;11(11):873–886.
- Shin SH, Bode AM, Dong ZG. Addressing the challenges of applying precision oncology. Npj Precis Oncol. 2017;1:28.
- Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219.
- Halait H, DeMartin K, Shah S, et al. Analytical performance of a real-time PCR-based assay for V600 mutations in the BRAF Gene, used as the companion diagnostic test for the Novel BRAF inhibitor vemurafenib in metastatic melanoma. Diagn Mol Pathol. 2012;21(1):1–8.
- Sapio MR, Posca D, Troncone G, et al. Detection of BRAF mutation in thyroid papillary carcinomas by mutant allele-specific PCR amplification (MASA). Eur J Endocrinol. 2006;154(2):341–348.
- Matsuda K. PCR-based detection methods for single-nucleotide polymorphism or mutation: real-time PCR and its substantial contribution toward technological refinement. Adv Clin Chem. 2017;80:45–72.
- Olmedillas-Lopez S, Garcia-Arranz M, Garcia-Olmo D. Current and emerging applications of droplet digital PCR in oncology. Mol Diagn Ther. 2017;21:493–510.
- Starostik P. Clinical mutation assay of tumors: new developments. Anti-Cancer Drugs. 2017;28(1):1–10.
- Kamps R, Brandao RD, Bosch BJ, et al. Next-Generation sequencing in oncology: genetic diagnosis, risk prediction and cancer classification. Int J Mol Sci. 2017;18(2):308.
- Yohe S, Thyagarajan B. Review of clinical next-generation sequencing. Arch Pathol Lab Med. 2017;141(11):1544–1557.
- Wu CC, Ko FH, Yang YS, et al. Label-free biosensing of a gene mutation using a silicon nanowire field-effect transistor. Biosens Bioelectron. 2009;25(4):820–825.
- Fang Z, Kelley SO. Direct electrocatalytic mRNA detection using PNA-nanowire sensors. Anal Chem. 2009;81(2):612–617.
- Huber F, Lang HP, Backmann N, et al. Direct detection of a BRAF mutation in total RNA from melanoma cells using cantilever arrays. Nat Nanotechnol. 2013;8(2):125–129.
- Wang Y, Tian K, Shi R, et al. Nanolock-Nanopore facilitated digital diagnostics of cancer driver mutation in tumor tissue. ACS Sensors. 2017;2(7):975–981.
- Zhang X, Price NE, Fang X, et al. Characterization of interstrand DNA-DNA cross-links using the alpha-hemolysin protein nanopore. ACS Nano. 2015;9(12):11812–11819.
- Nejad MI, Shi R, Zhang X, et al. Sequence-Specific covalent capture coupled with high-contrast nanopore detection of a disease-derived nucleic acid sequence. ChemBioChem. 2017;18(14):1383–1386.
- Villanueva MT. Melanoma: blocking BRAF to the BRIM. Nat Rev Clin Oncol. 2014;11(4):179.
- Ono A, Torigoe H, Tanaka Y, et al. Binding of metal ions by pyrimidine base pairs in DNA duplexes. Chem Soc Rev. 2011;40(12):5855–5866.
- Granzhan A, Kotera N, Teulade-Fichou MP. Finding needles in a basestack: recognition of mismatched base pairs in DNA by small molecules. Chem Soc Rev. 2014;43(10):3630–3665.
- Sato Y, Honjo A, Ishikawa D, et al. Fluorescent trimethyl-substituted naphthyridine as a ligand for C-C mismatch detection in CCG trinucleotide repeats. Chem Commun (Camb). 2011;47(20):5885–5887.
- Zeglis BM, Barton JK. DNA base mismatch detection with bulky rhodium intercalators: synthesis and applications. Nat Protoc. 2007;2(2):357–371.
- Lu CH, Willner I. Stimuli-responsive DNA-functionalized nano-/microcontainers for switchable and controlled release. Angewandte Chemie. 2015;54(42):12212–12235.
- Miyake Y, Togashi H, Tashiro M, et al. MercuryII-mediated formation of thymine-HgII-thymine base pairs in DNA duplexes. J Am Chem Soc. 2006;128(7):2172–2173.
- Kumar K, Isa L, Egner A, et al. Formation of nanopore-spanning lipid bilayers through liposome fusion. Langmuir. 2011;27(17):10920–10928.
- Wang G, Zhao Q, Kang X, et al. Probing mercury(II)-DNA interactions by nanopore stochastic sensing. J Phys Chem B. 2013;117(17):4763–4769.
- Torigoe H, Miyakawa Y, Ono A, et al. Thermodynamic properties of the specific binding between Ag+ ions and C:C mismatched base pairs in duplex DNA. Nucleosides Nucleotides Nucleic Acids. 2011;30(2):149–167.
- Price NE, Johnson KM, Wang J, et al. Interstrand DNA-DNA cross-link formation between adenine residues and abasic sites in duplex DNA. J Am Chem Soc. 2014;136(9):3483–3490.
- Cherf GM, Lieberman KR, Rashid H, et al. Automated forward and reverse ratcheting of DNA in a nanopore at 5-A precision. Nat Biotechnol. 2012;30(4):344–348.
- Kasianowicz JJ, Brandin E, Branton D, et al. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci U S A. 1996;93(24):13770–13773.
- Manrao EA, Derrington IM, Laszlo AH, et al. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat Biotechnol. 2012;30(4):349–U174.
- An N, Fleming AM, Middleton EG, et al. Single-molecule investigation of G-quadruplex folds of the human telomere sequence in a protein nanocavity. Proc Natl Acad Sci U S A. 2014;111(40):14325–14331.
- Cao C, Ying YL, Hu ZL, et al. Discrimination of oligonucleotides of different lengths with a wild-type aerolysin nanopore. Nat Nanotechnol. 2016;11(8):713–718.
- An N, Fleming AM, White HS, et al. Crown ether-electrolyte interactions permit nanopore detection of individual DNA abasic sites in single molecules. Proc Natl Acad Sci U S A. 2012;109(29):11504–11509.
- Wallace EV, Stoddart D, Heron AJ, et al. Identification of epigenetic DNA modifications with a protein nanopore. Chem Commun (Camb). 2010;46(43):8195–8197.
- Wang Y, Zheng D, Tan Q, et al. Nanopore-based detection of circulating microRNAs in lung cancer patients. Nat Nanotechnol. 2011;6(10):668–674.
- Wang Y, Luan BQ, Yang Z, et al. Single molecule investigation of Ag+ interactions with single cytosine-, methylcytosine- and hydroxymethylcytosine-cytosine mismatches in a nanopore. Sci Rep. 2014;4:5883.
- Rosen CB, Rodriguez-Larrea D, Bayley H. Single-molecule site-specific detection of protein phosphorylation with a nanopore. Nat Biotechnol. 2014;32(2):179–181.
- Wolfe AJ, Mohammad MM, Cheley S, et al. Catalyzing the translocation of polypeptides through attractive interactions. J Am Chem Soc. 2007;129(45):14034–14041.
- Wang S, Haque F, Rychahou PG, et al. Engineered nanopore of Phi29 DNA-packaging motor for real-time detection of single colon cancer specific antibody in serum. ACS Nano. 2013;7(11):9814–9822.
- Hornblower B, Coombs A, Whitaker RD, et al. Single-molecule analysis of DNA-protein complexes using nanopores. Nat Methods. 2007;4(4):315–317.
- Purnell RF, Schmidt JJ. Discrimination of single base substitutions in a DNA strand immobilized in a biological nanopore. ACS Nano. 2009;3(9):2533–2538.
- Kong J, Zhu J, Keyser UF. Single molecule based SNP detection using designed DNA carriers and solid-state nanopores. Chem Commun (Camb). 2016;53(2):436–439.
- Vercoutere W, Winters-Hilt S, Olsen H, et al. Rapid discrimination among individual DNA hairpin molecules at single-nucleotide resolution using an ion channel. Nat Biotechnol. 2001;19(3):248–252.
- Ang YS, Yung LY. Rapid and label-free single-nucleotide discrimination via an integrative nanoparticle-nanopore approach. ACS Nano. 2012;6(10):8815–8823.
- Cornelis S, Gansemans Y, Deleye L, et al. Forensic SNP genotyping using nanopore MinION sequencing. Sci Rep. 2017;7:41759.
- Zhao Q, Sigalov G, Dimitrov V, et al. Detecting SNPs using a synthetic nanopore. Nano Lett. 2007;7(6):1680–1685.
- Torigoe H, Miyakawa Y, Kozasa T, et al. The specific interaction between two T:T mismatch base pairs and mercury (II) cation. Nucleic Acids Symp Ser. 2007;51(1):185–186.
- Schmidt K, Mwaigwisya S, Crossman LC, et al. Identification of bacterial pathogens and antimicrobial resistance directly from clinical urines by nanopore-based metagenomic sequencing. J Antimicrob Chemother. 2017;72(1):104–114.
- Schmidt K, Mwaigwisya S, Crossman LC, et al. Identification of bacterial pathogens and antimicrobial resistance directly from clinical urines by nanopore-based metagenomic sequencing. J Antimicrob Chemother. 2017;72(1):104–114.
- Greninger AL, Naccache SN, Federman S, et al. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med. 2015;7:99.
- Jain M, Olsen HE, Paten B, et al. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol. 2016;17(1):239.
- Speicher MR, Pantel K. Tumor signatures in the blood. Nat Biotechnol. 2014;32(5):441–443.
- Chu D, Park BH. Liquid biopsy: unlocking the potentials of cell-free DNA. Virchows Arch. 2017;471:147–154.
- Pi C, Zhang MF, Peng XX, et al. Liquid biopsy in non-small cell lung cancer: a key role in the future of personalized medicine? Expert Rev Mol Diagn. 2017;17:1089–1096.
