The World Health Organization (WHO) reports that emerging and reemerging diseases worldwide are estimated to be responsible for millions of deaths each year. Due to the rapid spread and increasing number of infectious agents that continue to cause illness in the infected human being, simple, fast, inexpensive, sensitive, and specific diagnostic methods are necessary [Citation1]. Timely and accurate diagnostic approaches are crucial steps in controlling infectious diseases. The ongoing coronavirus disease 2019 (COVID-19) pandemic has proved the importance of developing new effective tests and making them available for rapid work-up and diagnosis. The polymerase chain reaction (PCR)-based methods for detecting microbial agent nucleic acid are the gold standard for diagnosing several infectious diseases. This method also applies to detect the causative agent of COVID-19, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [Citation2]. Such methods have limitations, including time-consuming, high cost, and the need for trained technicians. Another critical disadvantage is the risk of eliciting false-negative and false-positive results [Citation3]. Thus, there is a need to develop novel methods and improve technology with more rapid, sensitive, specific, and cost-effective methods.
Among different novel diagnostic methods, clustered regulatory, interspaced, short palindromic repeat (CRISPR), zinc-finger nucleases (ZFN), and transcription activator-like effector nuclease (TALEN) approaches are promising due to their programmability and ability to detect a specific location in the nucleic acid, i.e. site-specificity [Citation4]. CRISPR is widely used in functional gene screening, cell-based human hereditary disease modeling, epigenetic studies, and visualization of cellular processes [Citation5]. It is known as a rapid technology that quickly amplifies nucleic acid, with extremely high specificity and sensitivity. This assay is simple, efficient, and reliable and can distinguish differences in sequences of genomic targets. CRISPR gene editing is possible by ‘molecular scissors’ for cutting DNA threads that alter a piece of DNA. For example, new CRISPR-based technologies allow specific, multiplexed, portable, and ultrasensitive detection of RNA and DNA from clinically relevant samples [Citation6]. An important approach toward developing CRISPR-based molecular diagnostics was discovering the collateral activity of Cas12 and Cas13 proteins [Citation7]. A new version of CRISPR was introduced, called specific high-sensitivity enzymatic reporter unlocking (SHERLOCK), that incorporates genomic DNA strands with the CRISPR/Cas system to identify the targeted sequences [Citation6]. Another CRISPR-based diagnostic tool is the DNA endonuclease-targeted CRISPR trans reporter (DETECTOR), a rapid (∼30 min), low-cost, and accurate CRISPR-Cas12-based lateral flow assay for diagnosis of viral infections [Citation7].
Compared to other methods, CRISPR-based approaches are visual and a faster assay for molecular diagnosis. Importantly, studies reported 95% sensitivity and 100% specificity for this method [Citation6,Citation7]. In addition, there are studies that show prevention and treatment of acquired immunodeficiency syndrome (AIDS) by barring the entry of human immunodeficiency virus (HIV) into the cells or eliminating the integrated HIV genome from the human genome using CRISPR-Cas9 [Citation8]. However, this powerful technique has limitations due to the risk of causing other mutations, and undesirable effects like transferring unwelcomed genetic variations to future generations [Citation8]. Recently, studies have reported that the TALEN method is more efficient than CRISPR-Cas in heterochromatin parts of the genome [Citation9,Citation10]. In this method, transcription activator-like effectors (TALEs) are fused to a DNA scissor (Fok1) located in a spacer region, which facilitates genetic engineering. On TALEs, there are highly variable positions that act as guides to identify target nucleotides for binding [Citation11]. The DNA-binding domain of TALEs comprises monomers capable of binding to nucleotides in the target sequence. The TALEs DNA-binding domain includes tandem repeats of 34 residues of amino acids, 2 of which are highly variable (repeat variable diresidues, RVDs) and located at positions 12 and 13. These RVDs are recognition arms of TALEN for specific nucleotides. Their sequence is degenerate, and RVD monomers can bind to different nucleotides with various bonding affinity. Thus, TALEN could be a potential tool allowing scientists to design different ranges for both length and the number of tandem repeats. TALE can also be designed to identify a thymidine in the 5’ position of the target DNA. Therefore, before the TALE monomer attaches to a DNA, the 5’ of the target side should be thymine, which affects the attachment efficacy. TALEN is an innovative versatile tool to design various RVD arrays for gene editing and other applications such as diagnosis [Citation12].
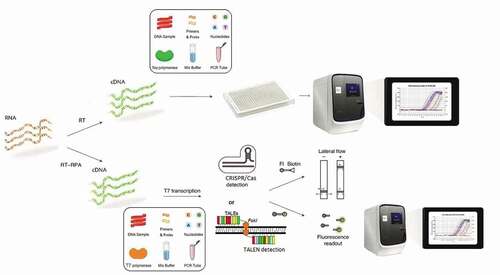
Increasing risks of false-positive results due to off-target activity and misidentification are very likely for some viruses sharing highly conserved sequences. For instance, human coronaviruses have identical structural genes, and it is difficult to distinguish between different viruses even with PCR [Citation3]. Moreover, SARS-CoV-2 variants may escape detection if sequence variation is within the CRISPR-Cas target region. A possible way to circumvent this problem is to develop multiplex CRISPR-Cas to detect more targets of the viral genome within the same or through parallel runs. On the other hand, off-targeting for TALEN seems to be lower than CRISPR, and TALEN often does not tolerate mismatches. A study showed that TALEN activity has no off-target in the human-induced pluripotent stem (iPS) cells [Citation13]. The procedure of TALEN is similar to CRISPR, albeit with more efficiency (). Briefly, the fragment of interest region in the RNA target under isothermal condition and by reverse transcription-recombinase polymerase amplification (RT-RPA) is amplified to DNA and then by T7 transcription converted to RNA. In CRISPR, the attachment of Cas13a-CRISPR RNA to amplified target RNAs stimulates the collateral activity of Cas13a, which then cleaves reporters. In TALEN, the same process is performed by TALEs and Fok1 endonuclease. Finally, separated RNA reporters should be captured on a colorimetric lateral-flow strip (biotin-fluorescein RNA reporter) or visualized by fluorescence signal (fluorescent reporter) [Citation14].
Figure 1. Illustration of workflow in Real-time PCR, CRISPR, and TALEN. In Real-time PCR, the trained operators require specific kits and facilities, but in CRISPR, a fragment of interest region of RNA target in isothermal condition and by reverse transcription-recombinase polymerase amplification (RT-RPA), is amplified to DNA and then by T7 transcription converted to RNA. The attachment of Cas13a–CRISPR RNA to amplified target RNAs stimulates the collateral activity of Cas13a, which then cleaves reporters. Separated RNA reporters should be captured on a colorimetric lateral-flow strip (biotin–fluorescein RNA reporter) or visualized by fluorescence signal (fluorescent reporter). A similar procedure is performed in TALEN but with more efficiency.
The sensitivity of CRISPR for virus detection is similar to approved point-of-care diagnostic assays like Cepheid Xpert Xpress and Abbott ID Now, and other assays under development that do not need particular facilities. This indicates that CRISPR’s suitability may be developed for application at site-of-care or at-home tests, performed where the individual is located. This may be less expensive than today’s work-up diagnostics, not pending the application of a safe sample-collection process. An at-home assay with broad coverage of infective Ct ranges can be valuable. CRISPR (SHERLOCK) can now detect SARS-CoV-2 in patients with Ct fewer than 33.5 by 100% specificity and 97–100% sensitivity. On the other hand, previous studies suggested more efficiency for TALEN [Citation9,Citation10]. These dynamic studies suggest that TALE could be used more efficiently in a more intimate heterochromatin environment than dCas9. Although CRISPR is a pioneer gene-editing approach, some critical challenges, such as off-target activity, remain.
The capability of the TALEN technique in engineering gene sequences will impact biological research and provide conditions to yield potential therapeutic strategies for disease [Citation15]. While TALEN has disadvantages such as a difficult protein engineering process that may cause uncontrolled mutations and disability to cleavage methylated DNA, the above approach with high site-specificity and flexibility must be considered as a potent approach in diagnosing infectious diseases. Although the Real-time PCR (RT-PCR) is critical for present COVID-19 management, the development of new novel methods will be useful and welcomed. Indeed, with the global threats of merging and reemerging viruses, the development of new approaches such as CRISPR or TALEN will be useful for rapid, specific, and on-site diagnostic work-up.
1. Expert opinion
TALEN has many advantages, such as unlimited target sites and high specificity. Compared to other methods, it has shown promising results and potential, and serves as an essential genome editing tool for diagnostic work-up and treatment issues in infectious diseases. The characteristics defined for this technique are promising, i.e. potential for rapid, on-site, and non-expensive diagnostic work-up, especially during pandemics. Although no study has been published so far using the TALEN method in COVID-19, this technique may be one of the most promising potential candidates to confirm COVID-19 disease.
Declaration of interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial relationships or otherwise to disclose.
Additional information
Funding
References
- Teymoori-Rad M, Samadizadeh S, Tabarraei A, et al. Ten challenging questions about SARS-CoV-2 and COVID-19. Expert review of respiratory medicine. Expert Rev Respir Med. 2020;14(9):881–888.
- Kohmer N, Rabenau HF, Hoehl S, et al. Comparative analysis of point-of-care, high-throughput and laboratory-developed SARS-CoV-2 nucleic acid amplification tests (NATs). J Virol Methods. 2021;291:114102.
- Tahamtan A, Ardebili A. Real-timeRT-PCR in COVID-19 detection: issues affecting the results. Expert review of molecular diagnostics. Expert Rev Mol Diagn. 2020;20(5):453–454.
- Sasano Y, et al. CRISPR-PCS: a powerful new approach to inducing multiple chromosome splitting in Saccharomyces cerevisiae. Sci. Rep. 2016;6(1):1–11.
- Mohammadinejad R, Biagioni A, Arunkumar G, et al. EMT signaling: potential contribution of CRISPR/Cas gene editing. Cell Mol Life Sci: CMLS. 2020;77(14):2701–2722.
- Kellner MJ, Koob JG, Gootenberg JS, et al. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Protoc. 2019;14(10):2986–3012.
- Broughton JP, et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38(7):870–874.
- Ayanoğlu FB, Elçin AE, Elçin YM. Bioethical issues in genome editing by CRISPR-Cas9 technology. Turk J Biol. 2020;44(2):110–120.
- Jain S, Shukla S, Yang C, et al. TALEN outperforms Cas9 in editing heterochromatin target sites. Nat Commun. 2021;12(1):1–10.
- Zhang S, Wang J, Wang J. One-Day TALEN assembly protocol and a dual-tagging system for genome editing. ACS omega. 2020;5(31):19702–19714.
- Malzahn AA, Qi Y. Assembly of TALEN and mTALE-act for plant genome engineering. Methods Mol Biol. 2021;2264:207–218. doi:https://doi.org/10.1007/978-1-0716-1201-9_15.
- Lamb BM, Mercer AC, Barbas CF III. Directed evolution of the TALE N-terminal domain for recognition of all 5′ bases. Nucleic Acids Res. 2013;41(21):9779–9785.
- Nemudryi A, Valetdinova KR, Medvedev SP, et al. TALEN and CRISPR/Cas genome editing systems: tools of discovery. Acta Naturae (англоязычная версия. Acta naturae. 2014;6(3):19–40.
- Takayama K, Igai K, Hagihara Y, et al. Highly efficient biallelic genome editing of human ES/iPS cells using a CRISPR/Cas9 or TALEN system. Nucleic Acids Res. 2017;45(9):5198–5207.
- Henderson E. TALEN is five times more efficient than CRISPR-Cas9 in tightly packed DNA; 2021. Available from: https://www.news-medical.net/news/20210127/Study-TALEN-is-five-times-more-efficient-than-CRISPR-Cas9-in-tightly-packed-DNA.aspx
