1. Introduction
The usual reaction to a sting from an insect of the order Hymenoptera is an inflammatory reaction that manifests in pain, swelling, erythema, and pruritus. In contrast, in Hymenoptera venom-allergic individuals, a single sting can lead to severe and even fatal anaphylaxis [Citation1]. The prevalence of insect venom allergy in the general population ranges from 0.3% to 7.5% [Citation2,Citation3]. In addition, this type I allergy, mediated by antibodies of the IgE isotype, is the most common trigger of severe anaphylaxis in adults [Citation4].
This editorial focuses on the common elicitors of venom allergy, honeybees (Apis mellifera) and yellow jackets (Vespula spp., known as yellow jackets in the US and wasps in Europe), for which molecular diagnostics of venom allergy is the most advanced [Citation5].
Patients at increased risk of systemic reactions to insect stings can be effectively treated by venom-specific immunotherapy (VIT), in which the allergy-eliciting venom is injected subcutaneously at regular intervals over several years after an initial up-dosing phase. VIT is currently the only causative, immunomodulatory therapy for venom allergy and protects 77–96% of patients from a future severe sting reaction [Citation3].
For an effective, potentially life-saving VIT, the clear identification of the allergy-eliciting venom is imperative. This includes the patient’s clinical history of a systemic sting reaction and the detection of specific IgE (sIgE) sensitization to the respective venom using skin tests (prick and/or intradermal testing) and/or the detection of venom-sIgE antibodies in the patient’s serum. In particular, if the patient could not identify the allergy-relevant insect or in case of contractionary results between clinical history and diagnostic tests, a clear diagnosis with classical venom extract-based diagnostics can be difficult. Here, molecular allergy diagnostics offers several advantages to strengthen therapeutic decisions and contribute to precision medicine in Hymenoptera venom allergy.
2. Venom extract-based sIgE diagnostics
Hymenoptera venoms are complex mixtures of a variety of substances that together mediate the toxic effects. These include numerous proteins, which represent potential allergens. In classical in vitro diagnostics, aqueous venom extracts are used in immunoassays to detect serum IgE antibodies specific to the venom allergens in the extracts.
In everyday clinical practice, over 50% venom-allergic patients show double-positive sIgE test results with honeybee venom (HBV) and yellow jacket venom (YJV) [Citation6]. These patients may have a primary sensitization to both venoms and therefore react with severe allergic reactions to the stings of both insects. For these patients, VIT with both venoms is recommended. However, for many of the double-positive patients, only one of the two venoms is of clinical relevance, while the positive diagnostic result for the second is due to cross-reactivity. In these patients, sufficient VIT success is limited to the primary sensitizing venom. Therefore, a clear distinction between primary allergy and cross-reactivity is an important clinical necessity to avoid unnecessary treatment with both venoms.
HBV and YJV contain several homologous pairs of allergens that share a high degree of similarity in protein sequence and three-dimensional structure () [Citation7]. Accordingly, they also share identical IgE epitopes. Thus, if a patient is sensitized to an HBV allergen, it is likely that the sIgE antibodies will also bind to its counterpart of YJV, even though there is no primary YJV sensitization (and vice versa). It is important to mention that protein cross-reactivity can also be of clinical relevance, which is reflected, for example, in the facts that several patients primarily sensitized to YJV can develop severe anaphylaxis after a hornet sting and vice versa and that hornet-allergic patients can be adequately treated with YJV VIT [Citation1,Citation8]. However, it was suggested that in patients with ascertained primary hornet allergy, VIT with hornet venom would be more adequate [Citation9]. Less is known about clinically relevant cross-reactivity between HBV and YJV.
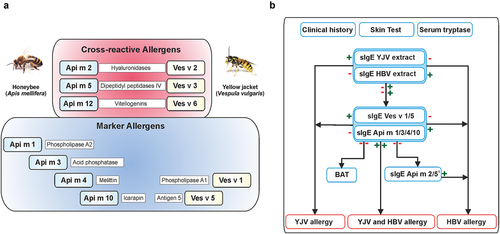
Figure 1. Molecular diagnostics of honeybee and yellow jacket venom allergy. A, Marker allergens and cross-reactive allergens of HBV and YJV venom. The allergen nomenclature is derived from the Latin species designations. In addition, the biochemical names of the allergens are given. B, Diagnostic algorithm for component-resolved diagnostics of HBV and YJV allergy. A ‘plus’ indicates a positive test result and a ‘minus’ indicates a negative test result. 1HBV allergens Api m 2 and Api m 5 show potential cross-reactivity to homologous allergens of YJV that are not commercially available, so a positive test result does not necessarily exclude YJV allergy. Despite the potential of component-resolved diagnostics, clinical history, skin tests, and the measurement of venom extract-sIgE and serum tryptase form an indispensable basis for an accurate diagnosis of Hymenoptera venom allergy. In addition, cellular tests such as basophil activation test (BAT) can be useful to dissect double-positive or double-negative test results. HBV, honeybee venom sIgE, specific IgE, YJV, yellow jacket venom.
In addition to this phenomenon, most HBV and YJV allergens carry cross-reactive carbohydrate determinants (CCDs), namely α-1,3-linked fucose at the innermost N-acetylglucosamine of the core structure of N-linked glycans, which are not found on human proteins. Therefore, a significant proportion of venom-allergic patients develops sIgE antibodies against these CCDs in addition to the allergen (protein)-specific ones. However, for reasons that have not yet been clarified, CCD-sIgE have no clinical relevance, i.e. they do not cause any allergic symptoms per se. However, since they bind to the CCDs of both HBV and YJV allergens, they also cause cross-reactivity in in vitro diagnostics [Citation10].
Both sIgE to homologous allergens and CCDs lead to cross-reactivities, which considerably complicates or even prevents the identification of the allergy-relevant venom using venom extracts for diagnostic purposes. In the past, this has often led to unnecessary treatments with HBV and YJV, since a clinically relevant allergy to both could not be ruled out.
In contrast to the scenario described above, there are patients in whom no sIgE sensitization is detected, despite a convincing history of anaphylaxis. Since evidence of sensitization is an essential prerequisite for initiating VIT, these patients can be excluded from this potentially life-saving treatment. Here, too the use of venom extracts for diagnostics can be the cause. Individual allergens may be under-represented or degraded in the extracts, their coupling efficiency to the solid phase of the assay can differ, or IgE epitopes can be masked by the coupling chemistry or natural binding partners in the extract.
Overall, venom extract-based diagnostics harbors several pitfalls that considerably complicate the identification of the allergy-eliciting venom and, thus, the selection of the optimal therapeutic strategy, particularly in patients who could not identify the allergy-relevant insect.
3. Molecular sIgE diagnostics of venom allergy
In recent years, increasing knowledge has been gained about the allergen composition of HBV and YJV, a fact that has shifted the perspective from the total venoms to individual allergens and has led to the development of molecular or component-resolved diagnostics (CRD) [Citation5]. Today, CRD is part of routine clinical diagnostics for venom allergy. With CRD, sIgE to individual allergens of the venoms are measured. In contrast to venom extract-based diagnostics, CRD not only provides information as to whether a patient has sIgE against the total venom, but also a sensitization profile at the molecular, allergen-resolved level. This implies that CRD can determine, which particular venom allergens are relevant for the individual patient.
In addition to homologous allergens, HBV and YJV also contain marker allergens that are only found in one of the two venoms (). Since CRD allows the determination of sIgE against these marker allergens as individual components, instead of being mixed with the cross-reactive allergens in a total venom extract, it has shown the potential to contribute significantly to the differentiation between relevant allergy and cross-reactivity [Citation7].
For CRD, the allergens are produced as recombinant proteins using host systems that allow their production with the complete protein epitope spectrum of the native allergens, but free of CCDs. In this way, allergens are obtained for which a positive sIgE test result only indicates true sensitization to the allergen and not also to its CCDs. Hence, recombinant CCD-free allergens additionally contribute to the distinction between primary allergy and cross-reactivity.
In addition, recombinant allergens are available in very pure form and almost unlimited quantities, so that CRD also circumvents the limitations of venom extract-based diagnostics, including under-representation, instability, inefficient coupling, or epitope masking.
Overall, CRD with recombinant CCD-free marker allergens provides an advanced tool for more precise diagnostics in many cases in which venom extract-based diagnostics does not allow clear identification of the allergy-relevant venom due to cross-reactivity or low sensitivity. Providentially, several of the relevant HBV and YJV allergens are available for CRD in clinical routine (). CRD with CCD-free single allergens is recommended: A) In the case of positive test results with different venoms, to discriminate between true sensitization and cross-reactivity. B) For diagnosis in patients with inconclusive anamnesis to identify the allergy-relevant insect(s). C) In the case of negative test results with various venoms despite a convincing clinical history due to potentially increased sensitivity of CRD.
Table 1. Characteristics of honeybee and yellow jacket venom allergens available for routine molecular diagnostics as of November 2022.
A diagnostic algorithm for distinguishing between HBV and YJV allergy, which considers the sIgE against individual allergens, is shown in (for more details on the diagnostics depicted see Blank et al. 2021 [Citation11]).
While CRD is able to adequately distinguish allergies to HBV and vespid venom (particularly YJV), this is not the case when differentiation between allergies to various vespid venoms is required. For instance, in Southern Europe, double-sensitization to YJV and European paper wasp (Polistes dominula) venom is very frequent. Here, definite discrimination is rarely possible due to the high degree of cross-reactivity between the major allergens of these venoms (for more details on this topic please refer to Blank et al. 2018 [Citation5] and Blank et al. 2021 [Citation7]).
4. Future potential of molecular diagnostics in venom allergy
Component-resolved analyses have demonstrated that allergic patients exhibit very individual sensitization profiles with the respective venom allergens [Citation12]. Recent work has shown that the success of VIT with HBV is associated with the type of allergen to which a patient is dominantly sensitized [Citation13]. In the case of a dominant sensitization (>50% of HBV-sIgE) to the allergen Api m 1, which is present in large amounts in the venom, there is no increased risk of treatment failure. On the other hand, if the patient is dominantly sensitized to the allergen Api m 10 [Citation14], which is only contained in traces in the venom, the risk of VIT failure increases more than eightfold. Accordingly, CRD could help to make predictive statements about the success of therapy and to improve therapeutic strategies, e.g. by choosing certain doses or preparations of venom for VIT [Citation15]. Sensitization to another HBV allergen (Api m 4) was demonstrated to be associated with a higher risk of side effects during the up-dosing phase of VIT [Citation16], which serves as further evidence that CRD could enable future optimization of treatment protocols.
The increasing availability of molecular diagnostics has opened up new options for resolving cross-reactivity and primary sensitization. In addition, it has shown potential for risk stratification in VIT and thus for personalized medical approaches in Hymenoptera venom allergy.
Declaration of interest
SB reports grants and personal fees from Bencard Allergie GmbH, grants and personal fees from Thermo Fisher Scientific, grants from LETI Pharma and Allergy Therapeutics, personal fees from the German Association for Allergology and Clinical Immunology (DGAKI), outside the submitted work. The other authors have nothing to disclose.
Reviewers disclosure
One reviewer discloses having received assay support from Thermo-Fisher/Phadia, but not for work relating to venom component-resolved diagnostics. The peer reviewers on this manuscript have no other relevant financial relationships or otherwise to disclose.
Additional information
Funding
References
- Blank S, Pehlivanli S, Methe H, et al. Fatal anaphylaxis following a hornet sting in a yellow jacket venom-sensitized patient with undetected monoclonal mast cell activation syndrome and without previous history of a systemic sting reaction. J Allergy Clin Immunol Pract. 2020;8(1):401–403 e402.
- Blank S, Haemmerle S, Jaeger T, et al. Prevalence of Hymenoptera venom allergy and sensitization in the population-representative German KORA cohort. Allergo J Int. 2019;28(6):183–191.
- Sturm GJ, Varga EM, Roberts G, et al. EAACI guidelines on allergen immunotherapy: hymenoptera venom allergy. Allergy. 2018;73(4):744–764.
- Worm M, Moneret-Vautrin A, Scherer K, et al. First European data from the network of severe allergic reactions (NORA). Allergy. 2014;69(10):1397–1404.
- Blank S, Bilo MB, Ollert M. Component-resolved diagnostics to direct in venom immunotherapy: important steps towards precision medicine. Clinical and Experimental Allergy: Journal of the British Society for Allergy and Clinical Immunology. 2018;48(4):354–364.
- Muller UR, Johansen N, Petersen AB, et al. Hymenoptera venom allergy: analysis of double positivity to honey bee and Vespula venom by estimation of IgE antibodies to species-specific major allergens Api m1 and Ves v5. Allergy. 2009;64(4):543–548.
- Blank S, Bilò MB, Grosch J, et al. Marker allergens in Hymenoptera venom allergy — characteristics and potential use in precision medicine. Allergo J Int. 2021;30(1):26–38.
- Kosnik M, Korosec P, Silar M, et al. Wasp venom is appropriate for immunotherapy of patients with allergic reaction to the European hornet sting. Croat Med J. 2002;43(1):25–27.
- Macchia D, Cortellini G, Mauro M, et al. Vespa crabro immunotherapy versus Vespula-venom immunotherapy in Vespa crabro allergy: a comparison study in field re-stings. World Allergy Organ J. 2018;11(1):3.
- Jappe U, Raulf-Heimsoth M, Hoffmann M, et al. In vitro hymenoptera venom allergy diagnosis: improved by screening for cross-reactive carbohydrate determinants and reciprocal inhibition. Allergy. 2006;61(10):1220–1229.
- Blank S, Grosch J, Ollert M, et al. Precision medicine in hymenoptera venom allergy: diagnostics, biomarkers, and therapy of different endotypes and phenotypes. Front Immunol. 2020;11:579409.
- Kohler J, Blank S, Muller S, et al. Component resolution reveals additional major allergens in patients with honeybee venom allergy. J Allergy Clin Immunol. 2014;133(5):1383–1389, 1389 e1381–1386.
- Frick M, Fischer J, Helbling A, et al. Predominant Api m 10 sensitization as risk factor for treatment failure in honey bee venom immunotherapy. J Allergy Clin Immunol. 2016;138(6):1663–1671 e1669.
- Jakob T, Rauber MM, Perez-Riverol A, et al. The honeybee venom major allergen Api m 10 (Icarapin) and its role in diagnostics and Treatment of hymenoptera venom allergy. Curr Allergy Asthma Rep. 2020;20(9):48.
- Blank S, Etzold S, Darsow U, et al. Component-resolved evaluation of the content of major allergens in therapeutic extracts for specific immunotherapy of honeybee venom allergy. Hum Vaccin Immunother. 2017;13(10):2482–2489.
- Ruiz B, Serrano P, Moreno C. IgE-Api m 4 is useful for identifying a particular phenotype of bee venom allergy. J Investig Allergol Clin Immunol. 2016;26(6):355–361.
