ABSTRACT
Introduction
Ulcerative colitis (UC) is a chronic illness requiring lifelong management that could be enhanced by personalizing care using biomarkers.
Areas covered
The main biomarker discovery modalities are reviewed, highlighting recent results across the spectrum of applications, including diagnostics (serum anti-αvβ6 antibodies achieving an area under the curve [AUC] = 0.99; serum oncostatin M AUC = 0.94), disease activity assessment (fecal calprotectin and serum trefoil factor 3: AUC > 0.90), prognostication of the need for treatment escalation (whole blood transcriptomic panels and CLEC5A/CDH2 ratio: AUC > 0.90), prediction of treatment response, and early identification of patients with subclinical disease. The use of established biomarkers is discussed, along with new evidence regarding autoantibodies, proteins, proteomic panels, transcriptomic signatures, deoxyribonucleic acid methylation patterns, and UC-specific glycomic and metabolic disturbances.
Expert opinion
Novel biomarkers will pave the way for optimized UC care. However, validation, simplification, and direct clinical translation of complex models may prove challenging. Currently, few candidates exist to assess key characteristics, such as UC susceptibility, histological disease activity, drug response, and long-term disease behavior. Further research will likely not only reveal new tools to tackle these issues but also contribute to understanding UC pathogenesis mechanisms.
1. Biomarkers in ulcerative colitis – hopes and challenges
Ulcerative colitis (UC) is a chronic inflammatory bowel disease (IBD) that affects 0.3% of the population of highly developed regions [Citation1] and is increasing in incidence in the developing world [Citation2]. Patients experience abdominal pain, bloody diarrhea, and fatigue, which lead to a substantial burden of disability, with approximately 10 million disability-adjusted life-years lost globally in the past decade alone [Citation3]. The relapsing and remitting course of UC necessitates life-long management involving repeated diagnostics and anti-inflammatory and immunosuppressive pharmacotherapy. A proportion of patients will require a proctocolectomy for acute severe colitis, medically-refractory disease, or cancer/dysplasia.
Gene–environment interactions, immune dysregulation, and dysbiosis are all known contributors to UC [Citation4], but despite large-scale epidemiology and multi-omics studies, the UC aetiopathogenesis picture is still incomplete. In parallel to unraveling the pathogenesis, a new focus has emerged in the UC community to leverage novel technologies for pragmatic biomarker discovery in clinical care that provide immediate patient benefit. Biomarker discovery to personalize care was selected as the second most important research theme in a James Lind Alliance survey [Citation5] and has also been listed as a key topic by a Crohn’s & Colitis UK survey, as highlighted in the recent British Society for Gastroenterology UC guidelines [Citation6].
The critical need to develop UC biomarkers has been and remains the common purpose of multiple research projects. However, these studies have great variations in methodology, study designs, and outcomes [Citation7] (). There are multiple contexts in which UC biomarkers could be used. They may facilitate diagnostics, distinguishing between UC and infectious gastroenteritis and UC and irritable bowel syndrome. Likewise, they could be used for disease monitoring by non-invasively determining endoscopic or histologic disease activity. Biomarkers could also be developed to prognosticate UC severity, predict treatment response, and monitor drug-related adverse events (). In addition, biomarkers could help to identify at-risk individuals and those who already present subclinical, potentially reversible pathology. They also have the potential to identify molecular UC subtypes and be employed to personalize UC care. Thus, UC biomarkers may have applications across a broad spectrum of clinical contexts and link to several different outcomes.
Figure 1. Biomarkers may support patient management throughout the disease course, from primary ulcerative colitis prevention to minimization of adverse event risk. The optimal selection of accurate biomarkers is the foundation of personalized medicine.
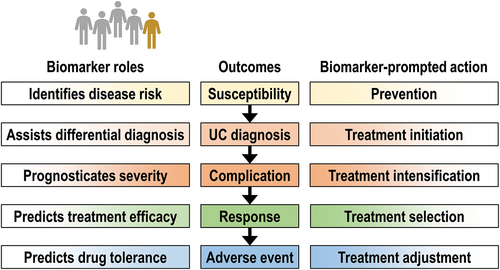
Table 1. The diversity of biomarker identification strategies in ulcerative colitis (UC) from the perspective of adoption and/or development in the near future.
Most immediately, biomarkers that reflect UC activity are required to reduce the burden of endoscopy and facilitate ‘treat-to-target’ algorithms, which is difficult to achieve without a consensus on a common histologic healing definition [Citation8], exposing a heterogeneity of outcomes. According to the CORE-IBD consensus initiative, another priority is the development of prognostication biomarkers [Citation9]. However, even definitions of treatment escalation differ, with some more inclusive and others focussing on strict clinical endpoints, such as surgery. As this framework of biomarker endpoints evolves [Citation10], much work is being undertaken using diverse laboratory and methodologic approaches. Here, we review these latest achievements in UC biomarker research, including the aforementioned variety of existing and proposed methods and the clinical needs that will guide further development in this area. We explore this landscape by listing laboratory methods, using examples to identify the most promising technologies, tackle fundamental questions, and propose solutions to facilitate discovery.
2. Established and investigational biomarkers in ulcerative colitis
2.1. The value of established ulcerative colitis biomarkers in current practice
2.1.1. Fecal calprotectin
Calprotectin is a dimer of S100A8 and S100A9 proteins that chelates zinc and iron, confers antibacterial activity [Citation11], and acts as an alarmin. It is one of the most abundant proteins in neutrophils and is released in the colon at the onset of inflammation. The high stability of calprotectin in feces makes it a convenient, noninvasive biomarker, and its diagnostic value is superior to the fecal immunochemical test [Citation12,Citation13]. Indeed, fecal hemoglobin was inferior to calprotectin in detecting mucosal healing (area under the receiver-operating characteristic curve [AUC] = 0.707 and 95% Confidence Interval (95%CI) = 0.620–0.784 vs. AUC = 0.858 and 95%CI = 0.784–0.913) (AUC 95%CI that extend above 0.5 implicate statistical significance of the findings) [Citation14]. A recent analysis of data from two large trials (GEMINI 1 and VARSITY) showed that post-induction fecal calprotectin (FC) of ≤250 μg/g predicted histological remission at one year (odds ratio [OR] = 5.54 and 95%CI = 3.77–8.14), and a lower risk of colectomy over seven years (hazard ratio [HR] = 0.30 and 95%CI = 0.13–0.68).
Since FC identifies histological remission in IBD [Citation15], its utility is embedded in national and international guidelines, including the British Society of Gastroenterology (BSG) [Citation5] and the European Society of Pediatric Gastroenterology, Hepatology and Nutrition (ESPGHAN)/European Crohn’s and Colitis Organization (ECCO) (pediatric) guidelines [Citation16]. As calprotectin was adopted worldwide, it became necessary to clarify the optimal scenarios for its use, with some societies releasing specific guidelines [Citation17,Citation18]. In general, calprotectin is advocated for the treat-to-target paradigm [Citation19], which is supported further by the fact that it correlates with patient-reported outcome measures [Citation20].
Fecal calprotectin use in clinical practice has certain limitations that need to be considered, including a relatively high normal upper limit in children and lower accuracy in proctitis [Citation21]. In general, FC is not UC or IBD-specific, highlighting the importance of understanding the appropriate context for its application. New developments suggest its role can extend to assessing biliary inflammation in primary sclerosing cholangitis (PSC) [Citation22], which often accompanies UC. Emerging data indicate poor uptake of fecal-based biomarkers amongst patients, with forgetfulness and sample collection commonly reported reasons for noncompliance [Citation23,Citation24]. As such, some IBD patients prefer serum biomarkers. Moreover, FC has a considerable variance, even within the same day [Citation25].
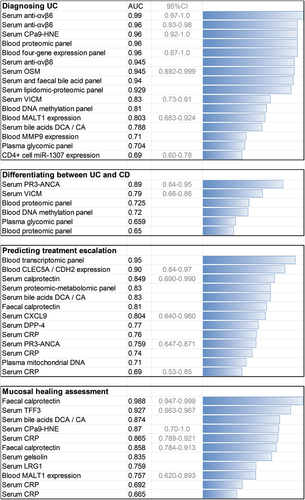
Although fecal assessment of calprotectin concentration is the most common marker, data suggest that additional benefits could be reached by assessing calprotectin in serum [Citation26] or plasma [Citation27]. Serum calprotectin provided the highest value in prognosticating flare (AUC = 0.849 and 95%CI = 0.690–0.990), followed by chemokine ligand 9 (CXCL9) (AUC = 0.804 and 95%CI = 0.640–0.960), in a study investigating 50 other putative biomarkers [Citation28] (). The calprotectin neo-epitope CPa9-HNE (elastase-cleaved calprotectin) can be used to measure neutrophil activity, which is different between UC patients and healthy controls (AUC = 0.96, 95%CI = 0.92–1.0, sensitivity = 88%, specificity = 100%, n = 66), and endoscopic remission vs. active disease (AUC = 0.87, 95%CI = 0.70–1, sensitivity = 80%, specificity = 94%) [Citation29]. Furthermore, low baseline CPa9-HNE predicted vedolizumab discontinuation in Crohn’s disease (CD) (AUC = 0.81, 95%CI 0.66–0.96, n = 33) [Citation30]. Therefore, the future may bring an appreciation of blood calprotectin, broaden the indications for its use, and hopefully harmonize laboratory methods and thresholds worldwide.
Figure 2. Comparison of the area under the receiver-operating characteristic curve (AUC) for some of the established and investigational ulcerative colitis biomarkers in various clinical contexts. The AUC values are presented to illustrate general trends in the field and not to compare individual studies, which strongly differ in design, e.g. a higher AUC is more easily achieved for short-term prognostication or when patients diagnosed with UC had more active disease. Only studies that reported an AUC are included, and in some cases, multiple studies provided data on the value of the same marker in the same application.
2.1.2. C-reactive protein
C-reactive protein (CRP) recognizes microbial and host lysolecithins to activate the complement system and is synthesized in the liver in response to interleukin-6 (IL-6). Although the latter cytokine can also be assessed in clinical settings, CRP is broadly available and has been better researched. Moreover, high CRP modestly correlates with endoscopic UC [Citation31,Citation32] and reflects UC clinical activity [Citation33]. There are conflicting results of direct comparisons against FC, with some studies suggesting an advantage for CRP [Citation34] and others indicating FC superiority [Citation35,Citation36]. CRP may predict response to steroids in children [Citation37], as well as to colectomy [Citation38], and is associated with patient response to infliximab [Citation38] and tofacitinib [Citation39]. However, there is uncertainty surrounding the influence of UC duration on the accuracy of CRP and the optimal threshold [Citation40,Citation41]. Similar to FC, CRP offers good sensitivity, but low specificity, by reflecting ongoing inflammation, and despite its value in short-term prognostication, there is insufficient data to show its association with long-term outcomes.
Understanding the limits of FC and CRP in UC is important for defining future aims in UC biomarker research (). Ideally, new markers should exhibit superiority over FC and CRP and/or provide information that the other two cannot yield. Research on UC biomarkers needs to extend beyond describing the intensity of inflammation by reflecting its specific characteristics or finer disturbances that underlie disease development and perpetuation.
2.2. Investigational biomarkers in ulcerative colitis
2.2.1. Individual antibodies and proteins
2.2.1.1. High proteinase 3 antineutrophil cytoplasmic antibodies are specific for UC diagnosis
Among the UC serological biomarkers, antineutrophil cytoplasmic antibodies (ANCA) have been used for many years and are associated with the diagnosis of UC vs. CD and PSC development. Recently, baseline high proteinase 3 ANCA (PR3-ANCA, a type of cytoplasmic ANCA) predicted the need for steroids at induction (AUC = 0.759 and 95%CI = 0.647–0.871; sensitivity and specificity of 75% and 69%, respectively, at ≥3.5 U/mL). It was not sensitive (44.5%) but was specific (95.6%) for UC at diagnosis and correlated with the Mayo score [Citation42]. These findings are in line with earlier results highlighting the high specificity of PR3-ANCA for UC vs. CD [Citation43], reaching an AUC = 0.89 (95%CI = 0.84–0.95) in some studies [Citation44]. However, the failure to replicate these results in Australia underscores the importance of methodological and other differences and the need for harmonization [Citation45]. Moreover, the distinction between UC and CD using PR3-ANCA is less marked in children [Citation46].
2.2.1.2. Anti-integrin αvβ6 antibodies are specific for ulcerative colitis and may identify subclinical disease
The latest developments in serological UC biomarkers include IgG class anti-integrin αvβ6 autoantibodies [Citation47], which readily distinguished between UC and IBS patients and healthy controls (AUC = 0.945), yielding a sensitivity of 76.3% and a specificity of 96.0% (OR = 77.1, 95%CI = 23.2–214, p < 0.0001) with a threshold set at the 95th percentile of absorbance in healthy controls. These findings successfully replicated the original study, which first reported sensitivity (92.0%) and specificity (94.8%) for UC diagnosis, at a threshold of mean + 3 SD of value in control sera [Citation48]. Anti-αvβ6 May predict UC development up to 10 years before onset [Citation49]. In the most recent study, the AUC for UC diagnosis using αvβ6 autoantibodies was 0.99 (95%CI = 0.97–1.0) and 0.96 (95%CI = 0.93–0.98) in two different cohorts (OR = 64.0 and 156.3). However, these antibodies were shown to possess only mild prognosticating value for adverse outcomes.
2.2.1.3. Calprotectin remains the most accurate fecal biomarker for ulcerative colitis diagnosis and prognostication
Fecal markers other than calprotectin are investigated, but it remains a benchmark measurement. Fecal myeloperoxidase correlated strongly positively with calprotectin (r = 0.82, p < 0.001), had similar value in assessing endoscopic disease activity, and performed as well as calprotectin in prognosticating treatment escalation (HR = 3.71, 95%CI 2.07–6.64, p < 0.01; >26 μg/g) [Citation50]. A fecal M2 isoform of pyruvate kinase was less useful than calprotectin in diagnosing UC [Citation51], while an increase in fecal eosinophil-derived neurotoxin preceded UC relapse [Citation52]. The eosinophil cationic protein was also investigated in IBD, where it positively correlated with IBD activity and showed potential value in patients with low FC [Citation53]. Likewise, fecal leukocyte esterase showed similar accuracy to FC in identifying IBD endoscopic activity (AUC = 0.80 vs. 0.85, respectively; cutoffs > 125 leukocytes/µL and >300 µg/g). Over 50% of 70 patients in the study had UC) [Citation54].
2.2.1.4. Serum trefoil factor 3 and serum amyloid a may reflect mucosal healing better than C-reactive protein
Leucine-rich alpha-2 glycoprotein (LRG1) performed similarly to CRP in identifying clinical remission and mucosal healing but was better at identifying deep remission and complete mucosal healing (AUC = 0.759 vs. 0.665 in a study involving 129 patients) [Citation55]. Another marker proposed for identifying complete mucosal healing is trefoil factor 3 (TFF3), which achieved an AUC = 0.927 (95%CI = 0.863–0.967) compared with 0.988 (95%CI = 0.947–0.999) for FC and 0.865 (95%CI = 0.789–0.921) for CRP [Citation56]. Serum amyloid A (SAA) identified patients scoring Mayo 2 or 3 better than CRP (AUC = 0.807, 95%CI = 0.748–0.867 vs. AUC = 0.701, 95%CI = 0.625–0.776; p = 0.01). However, SAA was not superior to CRP in patients with a UC duration of longer than 15 years [Citation57].
2.2.1.5. Dipeptidyl peptidase activity remains an investigational biomarker of mucosal healing
Dipeptidyl peptidase (DPP) activity was found to be potentially useful in IBD, although differences were modest, and only several UC patients were investigated. The AUC for DPP-4 in identifying UC activity vs. remission was 0.71 (95%CI = 0.60–0.82), lower than FC or serum CRP (AUC = 0.91 and 0.84, respectively). It was inferior to these established markers for describing endoscopic activity and had similar values for predicting treatment escalation (AUC for DPP-4, FC, and CRP: 0.77, 0.81, and 0.74, respectively) [Citation58,Citation59]. The use of multiple markers was associated with enhanced prognostication value [Citation59].
2.2.1.6. Serum gelsolin reflects mucosal healing better than C-reactive protein but is unspecific
Maeda et al. demonstrated in a retrospective, heterogeneous UC cohort (n = 138) that lower serum gelsolin could identify patients in clinical (AUC = 0.874; sensitivity and specificity of 78.41% and 86.54%) and endoscopic (AUC = 0.835) remission [Citation60]. These values were higher than for CRP (AUC = 0.780 and 0.692, respectively). However, changes in gelsolin concentrations have been described in multiple inflammatory disorders. Total globulin levels were found to be weakly associated with mucosal healing in patients with CRP <1 mg/L (AUC = 0.647; n = 147) [Citation61].
2.2.1.7. Serum oncostatin M is accurate in diagnosing ulcerative colitis and predicts anti-tumor necrosis factor non-response
Serum oncostatin M (OSM) was positively associated with a new diagnosis of UC vs. healthy controls (AUC = 0.945, 95%CI = 0.892–0.999), and accuracy was even higher when OSM or its receptor were measured in mucosal biopsies. In a mixed IBD cohort (with CD), mucosal OSM predicted non-response to anti-tumor necrosis factor (TNF) (AUC = 0.737, 95%CI = 0.537–0.937) and vedolizumab (AUC = 0.685, 95%CI = 0.529–0.840). Interestingly, in the same study, healthy first-degree relatives of multiple IBD patients also had elevated serum OSM [Citation62]. High baseline serum OSM predicted non-response to anti-TNF after one year and the need for rescue glucocorticosteroid use [Citation63]. Mucosal overexpression of OSM was found in infliximab-refractory IBD patients [Citation64]. OSM remains a promising biomarker, but its superiority over established biomarkers remains uncertain. IL-6, which is related to CRP and OSM, was associated with non-response to infliximab [Citation65], and its decrease correlated with vedolizumab efficacy [Citation66].
2.2.1.8. Collagen turnover reflects mucosal healing
Mortensen et al. investigated a set of promising collagen-related biomarkers, of which citrullinated and matrix metalloproteinase (MMP)-degraded vimentin (VICM) was most specific to UC, allowing for diagnosis (adjusted AUC = 0.83, 95%CI = 0.73–0.91) and distinction from CD (adjusted AUC = 0.79, 95%CI = 0.66–0.86) [Citation67]. A slight additional improvement in diagnostic value was achieved by adding degraded collagen III (C3M) to form a two-protein classifier. In another study, C3M alone could differentiate between UC remission and moderate or severe disease (AUC = 0.86, 95%CI = 0.54–0.98, sensitivity = 71%, specificity = 60%), and the diagnostic value increased after the addition of PRO-C3 (AUC = 0.80, 95%CI = 0.48–0.97, sensitivity = 86%, specificity 80%) [Citation68]. The ratio of C3M to PRO-C3 May be considered a measure of type III collagen turnover and interstitial matrix degradation by metalloproteinases.
2.2.2. Multiple proteins and clinical feature engineering
The use of a proximity extension assay (PEA) in an IBD-Character cohort enabled the discovery of a biomarker consisting of CD6 and colony-stimulating factor 1 (CSF1), which boosted the accuracy of predicting treatment escalation to 85.1% (95%CI = 79.2–91.0%) [Citation69], providing an improvement compared to CRP, especially in sensitivity. A rich PEA panel was also used by the Collaborative IBD Biomarker Research Initiative, which proposed an 8-protein (FGF19, signaling lymphocytic activation molecule family member 1 [SLAMF1], MMP10, IL10, IL17A, CSF1, IL12B, CX3CL1) biomarker panel to differentiate between UC and CD, with an AUC = 0.725 (n = 98) [Citation70]. Also, increased concentrations of six proteins (MMP10, CXCL9, SLAMF1, CXCL11, and monocyte chemoattractant protein [MCP]-1) were identified in subclinical UC by PEA and were elevated in healthy twins of patients relative to control [Citation71]. Analysis of over 1000 proteins, including antibodies, suggested that detection of subclinical UC is possible one year (AUC = 0.72) but not five years before clinical symptom onset (AUC = 0.56) (n = 199) [Citation72].
A combination biomarker for predicting endoscopic activity was proposed, consisting of SAA, IL-6, IL-8, and eotaxin-1, achieving an AUC = 0.84 (95%CI = 0.73–0.94), which was superior to CRP (AUC = 0.57, 95%CI = 0.43–0.72) in a mixed IBD cohort [Citation73]. Advanced mass spectrometry discovered proteomic UC markers in serum and urine [Citation74]. Several inflammation-related proteins (e.g. MMP10, CXCL9) were used to build diagnostic models for UC, which achieved an AUC = 0.96 in a replication cohort including 30 patients with UC, 30 with CD, and 30 healthy controls (AUC = 0.65 for UC vs. CD) [Citation75]. Aptamer-based proteomics has also been used to understand reactions to steroids and anti-TNF treatment and has potential in IBD biomarker discovery [Citation76].
Recent studies have gone beyond multi-marker protein models to integrate clinical parameters as composite markers, including clinical information. Most recently, Adams et al. described a simple prognosticating marker for steroid failure in acute severe UC (OR = 11.9, 95%CI = 10.8–13.0), which includes elevated CRP, low albumin, and high Ulcerative Colitis Endoscopic Index of Severity (UCEIS) [Citation77]. Earlier, a score encompassing CRP, albumin, and endoscopy showed an AUC = 0.754 for predicting steroid non-response in acute UC [Citation78]. The CRP/albumin ratio (CAR) at day three predicted response to steroid therapy in acute UC (AUC = 0.752) and could predict colectomy [Citation79]. CAR is also better associated with the Truelove-Witts score in acute severe UC compared with CRP alone [Citation80]. Also, high albumin to globulin ratio (>1.643) was associated with a Mayo endoscopic subscore of zero in patients with low CRP (OR = 4.38, 95%CI = 1.06–21.77) [Citation81].
2.2.3. Deoxyribonucleic acid methylation
There is much optimism related to the use of deoxyribonucleic acid (DNA) methylation analysis in generating predictive IBD biomarkers. IBD-Character built a model for predicting IBD treatment escalation using three methylation probes located at TAP1, RPTOR, and TESPA1, yielding an HR = 5.19 (95%CI = 2.14–12.56, p = 9.70 × 10−4) [Citation82]. Yet, analysis of CD8+ T cell epigenomes in children did not reveal any prognostic biomarkers in IBD [Citation83]. In CD, a two-loci diagnostic biomarker (VMP1 and RPS6KA2) was found (AUC = 0.98) [Citation84,Citation85], though this two-probe marker had less accuracy in UC (AUC = 0.73 and 0.71 for VMP1 and RPS6KA2 individually) [Citation86].
A new 12-methylation probe panel enabled the identification of UC and controls with an AUC = 0.81, and another set of probes (19 probes) distinguished between UC and CD (AUC = 0.72). A recent review underscored the heterogeneity of study methods and highlighted a set of 103 methylation probes consistently discriminating UC from controls [Citation87]. However, there are questions regarding methylation signature stability and the influence of changes in the proportions of cell types, both in the blood and in the intestinal epithelium [Citation88]. Nevertheless, simple sampling and development in analytical methods make methylation markers an exciting research thread.
2.2.4. Gene expression
One of the first blood gene expression profiling studies in UC showed that a four-gene panel (CD300A, KPNA4, IL1R2, and ELAVL1) enabled UC diagnosis (AUC = 0.96, 95%CI = 0.87–1.0) in an early demonstration of the potential of transcriptomics, which at the time required microarray analysis [Citation89]. More recently, a 17-gene biomarker using quantitative polymerase chain reaction (qPCR) and a regression model predicted a shorter time to treatment escalation in a UC validation cohort with an HR = 3.12 (95%CI = 1.25–7.72) [Citation90]. The test is already commercially available, and an ongoing trial will assess its value in CD (the PROFILE trial). In the IBD-Character cohort, a high two-gene ratio of CLEC5A to CDH2 expression (above 3) predicted the need for treatment escalation with an HR = 23.4 (95%CI = 5.3–102.0) (AUC = 0.90, 95%CI = 0.84–0.97, vs. CRP AUC = 0.69, 95%CI = 0.53–0.85, p = 0.016) [Citation91]. A CD4+ T cell microRNA-based marker for UC diagnosis was also identified (miR-1307: AUC = 0.69, 95%CI = 0.60–0.78) and still possessed diagnostic value after adjustment for CRP, albumin, and age [Citation92]. Moreover, miR-3615 predicted treatment escalation (HR = 2.55). MiRNAs attract much attention in IBD, and candidate markers are discussed in detail elsewhere [Citation93,Citation94].
There is a number of other issues related to the use of gene expression for diagnostics and prognostication in UC. Firstly, colonic biopsies may not be the optimal material for analyses, whereas blood provides useful information [Citation95]. A study by Ostrowski et al. illustrates this point well, as individual blood transcripts could be used for diagnosis of active pediatric UC (e.g. MMP9 AUC in qPCR validation = 0.97, n = 523 children) [Citation96]. However, this was dependent on ongoing inflammation. In another study, MALT1 expression in the blood identified active vs. quiescent UC (AUC = 0.757, 95%CI = 0.620–0.893) and quiescent UC vs. controls (AUC = 0.803, 95%CI = 0.683–0.924) [Citation97]. While individual transcripts may possess high diagnostic value, a polygenic transcriptomic risk score may offer additional advantage. Such score predicted colectomy with an AUC = 0.95, compared with an AUC = 0.74 for pediatric UC activity index (PUCAI) in a study involving 428 children [Citation98]. In summary, promising biomarkers, both simple and complex, have been identified though further validation is required.
2.2.5. Glycosylation
Analysis of plasma N-glycomes from over 1000 UC patients revealed differences in bisection, galactosylation, and sialylation compared with CD (AUC = 0.659) [Citation99]. Differences also existed in UC vs. controls (AUC = 0.704 in a replication cohort). A composite glycosylation biomarker (from the total serum N-glycome) was highly predictive of treatment escalation in UC (HR = 30.83, 95%CI = 7.9–120.4) [Citation100,Citation101]. Importantly, this universal IBD glycomic biomarker was validated in an external cohort. Furthermore, a low level of branched N-glycans in colon biopsies was associated with standard treatment non-response in patients with severe UC (AUC = 0.762, 95%CI = 0.571–0.953) [Citation102]. Despite the low availability of methods for glycomic marker discovery, they appear to have important translational potential.
2.2.6. Metabolomics
Implications of fatty acid metabolism and relatively frequent biliary involvement in UC point toward the potential value of metabolic biomarkers. Nuclear magnetic resonance of plasma revealed markers (lipoproteins and amino acids) reflecting high UC endoscopic activity (77% accuracy). Another metabolite profile in the same study was useful for prognostication over six months (74% accuracy, n = 60) [Citation103]. A different and arguably less expensive method, infrared spectroscopy, exhibited high sensitivity (100%) and specificity (86%) for UC diagnosis (n = 46) [Citation104]. Rectal positron emitting tomography (PET)-computed tomography (CT), which exposes the patient to considerable radiation, could be used for colitis severity assessment, but with less significance than FC, suggesting no broad clinical utility [Citation105].
A joint proteomic-metabolomic panel involving IL-10, carnitine, and sorbitol, among other factors, achieved an AUC = 0.83 for CD prediction and UC relapse (n = 164) [Citation106]. In an analysis of bile acids (n = 69 subjects), serum deoxycholic/cholic acid (DCA/CA) species had an AUC = 0.874 for differentiating between active UC and remission and an AUC = 0.788 for UC diagnosis vs. controls (compared with an AUC = 0.726 for CRP) [Citation107]. In the same study, more complex markers (analysis of sera and feces) yielded an AUC as high as 0.94. Serum bile acids were also found to associate with response to mesalazine [Citation108].
Fecal amino acids discriminated between children newly diagnosed with UC and controls (90% accuracy), and UC extent associated with tryptophan and other amino acids [Citation109]. High concentrations of taurine, homocitrulline, and kynurenine were more characteristic of CD than UC, providing a sensitivity of 88.4% and a specificity of 84.6% [Citation110].
Dyslipidaemia predicted the need for surgery (HR = 3.27, 95%CI = 1.86–5.75) and was associated with cancer development [Citation111]. An investigation of compound lipidome/proteome markers revealed that an AUC of 0.929 (sensitivity of 84.6% and specificity of 88.9%, n = 30) could be achieved for UC diagnosis [Citation112].
Even though the necessity to construct intricate panels of metabolites may appear as a limiting factor, such technologies are already in large-scale use in medicine, including newborn screening programs.
2.2.7. Microbiota
The involvement of microbiota in UC has attracted much attention, and expectations of its therapeutic potential are high, while data also indicate that microbiota might possess prognostic value. Increased proteolytic activity in stools precedes UC onset and is related to an altered functional profile of the fecal microbial communities [Citation113]. A random forest classifier trained on salivary microbiota (16S sequencing) allowed for discrimination between UC and PSC in children (AUC = 0.876, n = 48) [Citation114]. Also, in children, Ruminococcus gnavus abundance is positively associated with the extent of colitis [Citation115]. Microbiota-based indices were proposed for diagnosis, phenotype assessment, and prognostication, with higher Bifidobacteria abundance distinguishing UC from CD [Citation116]. In addition, more Bacteroides fragilis were found in the feces of UC patients than controls, and some species were associated with colitis extent (e.g. Bifidobacteria), but none predicted response to anti-TNF [Citation117]. In contrast, a high abundance of Verrucomicrobiota, together with two short-chain fatty acids, predicted rapid remission in response to vedolizumab (AUC = 0.961, 95%CI = 0.882–1.0) [Citation118].
Fecal microbiota transplantation success was predicted by high production of butyrate and Filobasidium spp [Citation119,Citation120]. Broad access to microbiota profiling is likely to facilitate further investigation and adoption of bacterial UC biomarkers. A large study is ongoing in the UK (IBD-RESPONSE) and seeks to recruit over 1000 patients with IBD to generate microbial biomarkers predictive of response to multiple therapies, including Janus kinase inhibitors.
2.2.8. Full blood count analysis
There have been attempts to exploit easily available data from complete blood counts as UC biomarkers. Neutrophil-to-lymphocyte ratio predicted mucosal healing in response to anti-TNF at week 54 [Citation66] and was weakly associated with the future development of pouchitis [Citation121]. The neutrophil-to-lymphocyte ratio also had a similar AUC to CRP (0.971; sensitivity, specificity, and positive predictive value – all at 90%; sample size n = 80) for the identification of active UC [Citation122]. In UC, a high neutrophil-to-lymphocyte ratio also associates with future relapse in patients with mucosal healing (HR = 1.74, 95%CI = 1.02–2.98) [Citation123]. Another interesting marker is monocytosis (exceeding the local upper limit of the norm), which predicts the need for hospitalization (HR = 6.5), even after adjustment for inflammatory markers (n = 1290) [Citation124]. In the era of biologics, analysis of basic leukocyte subtypes does not seem attractive. Nonetheless, it may be pragmatic, as demonstrated by the relationship between mucosal eosinophilia and the vedolizumab response described below.
2.2.9. Other research directions
There are several other research directions related to the discovery of UC biomarkers, ranging from polygenic risk scores [Citation125] to fluorescence-enhanced endoscopy [Citation126] and the assessment of retinal microcirculation using optical coherence tomography [Citation127]. New ultrasound scores have also been proposed (e.g. TIGER [Citation128], DUBLIN [Citation129]), and this type of noninvasive imaging may prove to be useful in prognostication, despite between-operator variability. Intestinal submucosal fibrosis associates with chronic inflammation and non-response to biologics [Citation130] and might serve to profile patients with UC [Citation131].
Mitochondrial DNA demonstrated similar performance to CRP in predicting colectomy in UC (AUC = 0.71 vs. 0.76, n = 67) [Citation132], and mitochondrial damage-associated molecular patterns are currently being investigated as CD biomarkers [Citation133]. There are also attempts to develop molecular inflammation scores to better reflect IBD activity in an understandable way, which could be linked with outcomes [Citation134].
Finally, incorporating the search for biomarkers in clinical trial design [Citation135] is poised to provide new insights, as exemplified by prediction of the vedolizumab response by mucosal eosinophil count (AUC = 0.90, n = 24) [Citation136], α4β7 positivity of CD3+ T cells in colonic biopsies (sensitivity and specificity of 100%; n = 21) [Citation137], and the association of the golimumab response with a transcriptomic signature in colonic biopsies (AUC = 0.671 at week 30; lower bound of 95%CI = 0.569) [Citation138].
2.3. The most promising biomarkers: a summary
Currently, the most promising biomarkers for identifying UC risk are anti-αvβ6 antibodies and stool proteolytic activity, which could be supplemented by a polygenic risk score. For UC diagnosis, anti-αvβ6 antibodies, PR3-ANCA levels, serum OSM, and serum CPa9-HNE could also be used. FC still has the best diagnostic value for mucosal healing assessment, but serum calprotectin is an interesting alternative, and several biomarkers are in development, such as serum TFF3, bile acids, CPa9-HNE, and gelsolin. Transcriptomic biomarkers (like CLEC5A/CDH2 or more complex panels) can strengthen prognostication, but calprotectin remains the most accurate of the clinically available tools. Two novel markers, anti-αvβ6 antibodies, and serum OSM, closest to adoption into practice, are useful in diagnosis but limited by their sensitivity.
3. Expert opinion
3.1. Recent progress
Over the last decade, there has been a huge emphasis on personalized care in CD and in UC, and discovering biomarker signatures in the blood, gut, or stool has become a critical focus in research and translational clinical care. The development of new technology for multi-omic analyses and machine learning for the analysis of complex datasets have added impetus to research. There are also exciting insights emerging in a host of clinical scenarios, including pre-clinical UC, early diagnosis, prognostication of clinical course, and response to drug therapy. However, no new biomarker has made the successful translation from research to widespread clinical practice. Many markers have shown exciting early data in discovery publications and have been the subject of patent applications and commercialization in a few cases. Considering the reasons underlying this failure to translate is worthwhile.
3.2. Major limitations and challenges
The most evident limitation holding back the translation of exciting early data on new markers for use in daily practice is the need for stringent, compelling, and unequivocal independent validation. Such studies can be difficult, costly, and take a long time to accomplish. Moreover, replication studies remain undervalued in an academic context and may be relatively difficult to fund or publish because of the perceived lack of scientific novelty. This remains an important area for discussion. Another challenge lies, paradoxically, in the rapid advances in technology, computing, and the associated plasticity and susceptibility to variation of complex models and machine learning algorithms that underlie diagnostic panels. The models may be sensitive to changes in laboratory methods or even individual reagents, which may escalate costs, reduce availability, and hamper adoption. Furthermore, any test developed for daily clinical practice needs to be simple, cheap, easy to use, rapid, highly specific for the disease of interest, and, most importantly, reproducible across different laboratories [Citation139].
3.3. Preparing the clinical implementation of biomarkers
Notwithstanding the highlighted caveats, the progress summarized in this review provides substantial grounds for optimism. Work within the field of transcriptomic biomarkers has perhaps defined the ideal path from biomarker discovery toward future clinical translation. While the initial studies identified a T-cell exhaustive gene expression signature in IBD, this qPCR panel was able to predict treatment escalation over time. Critical signal validation was then sought before the development of a commercial panel. Although there has been some controversy regarding the stability of this signal across independent cohorts [Citation83], the PROFILE trial was set up across UK sites and recently completed recruitment.
Most recently, in CD, the METHYLOMICS study has been initiated as a biomarker-driven study to personalize the use of biological therapies based on blood methylation profiles. In the future, biomarker-led clinical studies may become widely accepted, with potential markers discussed above, including anti-αvβ6 antibodies, PR3-ANCA, and VICM for UC diagnosis, SAA or OSM for activity assessment, and CLEC5A/CDH2 expression, transcriptomics, and glycomics for mid-term prognostication. Also, compound omics (e.g. lipidome/proteome) are now being used and yield promising results. Furthermore, there is evidence positioning microbiota characteristics or specific strains as UC biomarkers, but it will take large multifaceted prospective studies to add more certainty, especially given differences in sample handling and the inter- and intra-individual variability of bacterial communities. Some of these markers are likely to be broadly adopted into clinical practice within the next decade.
3.4. Towards personalized medicine in inflammatory bowel disease
There is clear heterogeneity in how we clinically define IBD, with several variations in presentation, sub-phenotype, and outcomes. With the approval of a number of new therapies for UC management, there is now a real unmet need to personalize drug therapies to provide tailored treatment with the greatest benefit and least amount of side effects for each patient. An example of such issue, which may benefit from the use of specific diagnostic tools is intestinal fibrosis, especially fibrostenosis, for which tailored biomarkers are in development, as mentioned above.
It may appear paradoxical that the field of biomarker research is so attractive because of disease heterogeneity and the incomplete efficacy of treatments. Even though it does not seem highly probable currently, the next decade might hypothetically bring a therapeutic breakthrough that would enable safe treatment with few side effects to almost all UC patients. Likewise, uncovering the cause of UC, and paving a way toward preventing the disease, could profoundly shift research priorities in the field.
Currently, a personalized approach based on biomarkers remains one of the most attractive topics in UC since it is poised to bring significant improvements to patients’ well-being in the near future. Set up and refinement of collaborative endeavors, such as the IBD Bioresource in the UK, are vital for providing the IBD research community with large well-phenotyped platforms for accessing clinical data and biosamples to help address key clinical scenarios and facilitate robust signal validation and subsequent clinical application.
Article highlights
Ulcerative colitis (UC) is a chronic, relapsing, and remitting inflammatory bowel disease that leads to a substantial burden of disability and affects 0.3% of the population of highly developed countries. There is a great need for personalized medicine in UC to optimize treatment and prevent complications.
Biomarkers are one of the priorities in UC research because of their potential to support diagnosis, noninvasive monitoring of endoscopic and histologic activity, prognostication of disease severity, and prediction of the response to treatment or drug-related adverse events.
Fecal calprotectin and serum C-reactive protein remain the most accessible UC activity biomarkers, but they offer limited specificity and insufficient mid to long-term prognostication accuracy.
According to recent research, anti-integrin αvβ6 autoantibodies and proteinase 3 antineutrophil cytoplasmic antibodies present excellent diagnostic value in UC. Moreover, anti-αvβ6 antibodies may enable subclinical screening in groups at high genetic risk of UC.
Attempts to refine the use of calprotectin through serum measurement or focus on the elastase-cleaved calprotectin neo-epitope CPa9-HNE are yielding promising results.
Other investigative biomarkers include serum trefoil factor 3, bile acids, gelsolin, stool proteolytic activity, and polygenic risk scores.
Transcriptomic, proteomic, metabolic, and glycomic panels can surpass other markers of UC severity prognostication accuracy, but there are challenges in clinical translation due to the complexity and sensitivity of predictive models.
Further progress in the field depends on improvements in infrastructure and collaboration and the alignment of definitions, outcomes, and research designs. The rapid adoption of the most performant and simple biomarkers is expected. Biomarker research will likely affect the understanding of UC pathogenesis.
Declaration of interests
JK Nowak reports a grant from Biocodex Microbiota Foundation, outside of the submitted work. R Kalla has served as a speaker for Ferring and has received support for research from IBD-Character [EU FP7 2858546]. J Satsangi has served as a speaker, a consultant, and an advisory board member for MSD, Ferring, Abbvie, and Shire, a consultant with Takeda, has received speaking fees from MSD, travel support from Shire, and has received research funding from Abbvie, Wellcome, CSO, MRC, and the EC grant IBDBIOM.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
Author contributions
JK Nowak – conceptualization, investigation, drafting the manuscript, funding acquisition, visualization; R Kalla – investigation, writing – original draft; J Satsangi – conceptualization, funding acquisition, investigation, project administration, supervision, writing – review & editing.
Additional information
Funding
References
- Ng SC, Shi HY, Hamidi N, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2017;390(10114):2769–2778.
- Agrawal M, Christensen HS, Bøgsted M, et al. The rising burden of inflammatory bowel disease in Denmark over two decades: a nationwide cohort study. Gastroenterology. 2022;163(6):1547–1554.e5.
- Alatab S, Sepanlou SG, Ikuta K, et al. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990–2017: a systematic analysis for the global burden of disease study 2017. Lancet Gastroenterol Hepatol. 2020;5(1):17–30.
- Graham DB, Xavier RJ. Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature. 2020;578(7796):527–539. doi: 10.1038/s41586-020-2025-2
- Lamb CA, Kennedy NA, Raine T, et al. British Society of Gastroenterology consensus guidelines on the management of inflammatory bowel disease in adults. Gut. 2019;68(Suppl 3):s1–s106.
- Zilbauer M, Heuschkel R. Disease prognostic biomarkers in inflammatory bowel diseases—A reality check. J Crohns Colitis. 2022;16(1):162–165. doi: 10.1093/ecco-jcc/jjab118
- Honig G, Heller C, Hurtado-Lorenzo A. Defining the path forward for biomarkers to address unmet needs in inflammatory bowel diseases. Inflamm Bowel Dis. 2020;26(10):1451–1462. doi: 10.1093/ibd/izaa210
- Honig G. Towards a common validated methodology for histologic measurement of mucosal healing in ulcerative colitis: introducing a new project of the biomarkers consortium of the foundation for the nih. Gastroenterology. 2022;162(3):S35. doi: 10.1053/j.gastro.2021.12.071
- Ma C, Hanzel J, Panaccione R, et al. CORE-IBD: a multidisciplinary international consensus initiative to develop a core outcome set for randomized controlled trials in inflammatory bowel disease. Gastroenterology. 2022;163(4):950–964.
- Plevris N, Lees CW. Disease monitoring in inflammatory bowel disease: evolving principles and possibilities. Gastroenterology. 2022;162(5):1456–1475.e1. doi: 10.1053/j.gastro.2022.01.024
- Jukic A, Bakiri L, Wagner EF, et al. Calprotectin: from biomarker to biological function. Gut. 2021;70(10):1978–1988.
- Udegbune M, Sharrod–Cole H, Townsend S, et al. Diagnostic performance of serum calprotectin in discriminating active from inactive ulcerative colitis in an outpatient setting. Ann Clin Biochem. 2022;59(6):404–409.
- Shimizu H, Ebana R, Kudo T, et al. Both fecal calprotectin and fecal immunochemical tests are useful in children with inflammatory bowel disease. J Gastroenterol. 2022;57(5):344–356.
- Kim ES, Lee HS, Kim SK, et al. Fecal calprotectin is more accurate than fecal immunochemical test for predicting mucosal healing in quiescent ulcerative colitis: a prospective multicenter study. Scand J Gastroenterol. 2020;55(2):163–168.
- Kawashima K, Oshima N, Kishimoto K, et al. Low fecal calprotectin predicts histological healing in patients with ulcerative colitis with endoscopic remission and leads to prolonged clinical remission. Inflamm Bowel Dis. 2023;29(3):359–366.
- Turner D, Ruemmele FM, Orlanski-Meyer E, et al. Management of paediatric ulcerative colitis, part 1: ambulatory care—an evidence-based Guideline from European Crohn’s and colitis organization and European Society of paediatric Gastroenterology, hepatology and nutrition. J Pediatr Gastroenterol Nutr. 2018;67(2):257–291.
- Guardiola J, Lobatón T, Cerrillo E, et al. Recomendaciones del Grupo Español de Trabajo en Enfermedad de Crohn y Colitis Ulcerosa (GETECCU) sobre la utilidad de la determinación de calprotectina fecal en la enfermedad inflamatoria intestinal. Gastroenterol Hepatol. 2018;41(8):514–529.
- Koninckx CR, Donat E, Benninga MA, et al. The use of fecal calprotectin testing in paediatric disorders: a position paper of the European Society for paediatric Gastroenterology and nutrition gastroenterology committee. J Pediatr Gastroenterol Nutr. 2021;72(4):617–640.
- Dulai PS, Feagan BG, Sands BE, et al. Prognostic value of fecal calprotectin to inform treat-to-target monitoring in ulcerative colitis. Clin Gastroenterol Hepatol. 2023;21(2):456–466.e7.
- Kamat N, Vuyyuru SK, Kedia S, et al. Correlation of fecal calprotectin and patient-reported outcome measures in patients with ulcerative colitis. Intest Res. 2022;20(2):269–273.
- Sakuraba A, Nemoto N, Hibi N, et al. Extent of disease affects the usefulness of fecal biomarkers in ulcerative colitis. BMC Gastroenterol. 2021;21(1):197.
- Pavlidis P, Joshi D, Sherif YE, et al. Faecal calprotectin is a surrogate marker of biliary inflammation in primary sclerosing cholangitis associated inflammatory bowel disease. Frontline Gastroenterol. 2022;13(6):497–502.
- Kalla R, Boyapati R, Vatn S, et al. Patients’ perceptions of faecal calprotectin testing in inflammatory bowel disease: results from a prospective multicentre patient-based survey. Scand J Gastroenterol. 2018;53(12):1437–1442.
- Maréchal C, Aimone-Gastin I, Baumann C, et al. Compliance with the faecal calprotectin test in patients with inflammatory bowel disease. United Eur Gastroenterol J. 2017;5(5):702–707.
- Lasson A, Stotzer P-O, Öhman L, et al. The intra-individual variability of faecal calprotectin: a prospective study in patients with active ulcerative colitis. J Crohns Colitis. 2015;9:26–32. doi: 10.1016/j.crohns.2014.06.002
- Kalla R, Kennedy NA, Ventham NT, et al. Serum Calprotectin: a Novel diagnostic and prognostic marker in inflammatory bowel diseases. Am J Gastroenterol. 2016;111(12):1796–1805.
- Malham M, Carlsen K, Riis L, et al. Plasma calprotectin is superior to serum calprotectin as a biomarker of intestinal inflammation in ulcerative colitis. Scand J Gastroenterol. 2019;54(10):1214–1219.
- Kessel C, Lavric M, Weinhage T, et al. Serum biomarkers confirming stable remission in inflammatory bowel disease. Sci Rep. 2021;11(1):6690.
- Mortensen JH, Sinkeviciute D, Manon-Jensen T, et al. A specific calprotectin neo-epitope [CPa9-HNE] in serum from inflammatory bowel disease patients is associated with neutrophil activity and endoscopic severity. J Crohns Colitis. 2022;16(9):1447–1460. doi: 10.1093/ecco-jcc/jjac047
- Alexdottir MS, Bourgonje AR, Karsdal MA, et al. Serological biomarkers of extracellular matrix turnover and neutrophil activity are associated with long-term use of vedolizumab in patients with Crohn’s disease. Int J Mol Sci. 2022;23(15):8137.
- Iwańczak B, Ruczka M, Matusiewicz M, et al. Correlation between biomarkers (calprotectin, seromucoid, metalloproteinase-3 and CRP) and clinical and endoscopic activity of ulcerative colitis in children. Adv Med Sci. 2020;65(2):259–264.
- Yoon JY, Park SJ, Hong SP, et al. Correlations of C-reactive protein levels and erythrocyte sedimentation rates with endoscopic activity indices in patients with ulcerative colitis. Dig Dis Sci. 2014;59(4):829–837.
- Karoui S, Laz S, Serghini M, et al. Correlation of C-reactive protein with clinical and endoscopic activity in patients with ulcerative colitis. Dig Dis Sci. 2011;56(6):1801–1805.
- Ishida N, Higuchi T, Miyazu T, et al. C-reactive protein is superior to fecal biomarkers for evaluating colon-wide active inflammation in ulcerative colitis. Sci Rep. 2021;11(1):12431.
- Voiosu T, Benguş A, Bălănescu P, et al. Rapid fecal calprotectin Testing predicts mucosal healing better than C-reactive protein and serum tumor necrosis factor α in patients with ulcerative colitis. Romanian J Intern Med Rev Roum Med Interne. 2015;53(3):253–260.
- Schoepfer AM, Beglinger C, Straumann A, et al. Fecal calprotectin more accurately reflects endoscopic activity of ulcerative colitis than the Lichtiger index, C-reactive protein, platelets, hemoglobin, and blood leukocytes. Inflamm Bowel Dis. 2013;19(2):332–341.
- Deva Rajoo G, Tan L, Lopez A, et al. Early response to corticosteroid and baseline C-Reactive protein predicts outcomes in children with moderate to severe ulcerative colitis. Dig Dis Sci. 2019;64(7):1929–1937.
- Con D, Andrew B, Nicolaides S, et al. Biomarker dynamics during infliximab salvage for acute severe ulcerative colitis: C-reactive protein (CRP)-lymphocyte ratio and CRP-albumin ratio are useful in predicting colectomy. Intest Res. 2022;20(1):101–113.
- Dubinsky MC, Magro F, Steinwurz F, et al. Association of C-reactive protein and partial Mayo score with response to tofacitinib induction therapy: results from the ulcerative colitis clinical program. Inflamm Bowel Dis. 2022;29(1):51–61.
- Croft A, Lord A, Radford-Smith G. Markers of systemic inflammation in acute attacks of ulcerative colitis: what level of C-reactive protein constitutes severe colitis? J Crohns Colitis. 2022;16(7):1089–1096. doi: 10.1093/ecco-jcc/jjac014
- Bakkaloglu OK, Eskazan T, Celik S, et al. Can we predict mucosal remission in ulcerative colitis more precisely with a redefined cutoff level of C-reactive protein? Colorectal Dis Off J Assoc Coloproctology G B Irel. 2022;24(1):77–84.
- Imakiire S, Takedatsu H, Mitsuyama K, et al. Role of serum proteinase 3 antineutrophil cytoplasmic antibodies in the diagnosis, evaluation of disease severity, and clinical course of ulcerative colitis. Gut Liver. 2022;16(1):92–100.
- Horn MP, Peter AM, Righini Grunder F, et al. PR3-ANCA and panel diagnostics in pediatric inflammatory bowel disease to distinguish ulcerative colitis from Crohn’s disease. PLoS One. 2018;13:e0208974. doi: 10.1371/journal.pone.0208974
- Xu Y, Xu F, Li W, et al. The diagnostic role and clinical association of serum proteinase 3 anti-neutrophil cytoplasmic antibodies in Chinese patients with inflammatory bowel disease. Scand J Gastroenterol. 2020;55(7):806–813.
- Lee W-I, Subramaniam K, Hawkins CA, et al. The significance of ANCA positivity in patients with inflammatory bowel disease. Pathology. 2019;51(6):634–639.
- Laass MW, Ziesmann J, de Laffolie J, et al. Anti-proteinase 3 antibodies as a biomarker for ulcerative colitis and primary sclerosing cholangitis in children. J Pediatr Gastroenterol Nutr. 2022;74(4):463–470.
- Rydell N, Ekoff H, Hellström PM, et al. Measurement of serum IgG anti-integrin αvβ6 autoantibodies is a promising tool in the diagnosis of ulcerative colitis. J Clin Med. 2022;11(7):1881.
- Kuwada T, Shiokawa M, Kodama Y, et al. Identification of an anti–integrin αvβ6 autoantibody in patients with ulcerative colitis. Gastroenterology. 2021;160(7):2383–2394.e21.
- Livanos AE, Dunn A, Fischer J, et al. Anti-Integrin αvβ6 Autoantibodies Are a Novel Biomarker That Antedate Ulcerative Colitis. Gastroenterology. 2023;164(4):619–629. doi: 10.1053/j.gastro.2022.12.042
- Swaminathan A, Borichevsky GM, Edwards TS, et al. Faecal myeloperoxidase as a biomarker of endoscopic activity in inflammatory bowel disease. J Crohns Colitis. 2022;16(12):1862–1873.
- Czub E, Nowak JK, Szaflarska-Poplawska A, et al. Comparison of fecal pyruvate kinase isoform M2 and calprotectin in assessment of pediatric inflammatory bowel disease severity and activity. Acta Biochim Pol. 2014;61(1):99–102.
- Amcoff K, Cao Y, Zhulina Y, et al. Prognostic significance of faecal eosinophil granule proteins in inflammatory bowel disease. Scand J Gastroenterol. 2019;54(10):1237–1244.
- Abedin N, Seemann T, Kleinfeld S, et al. Fecal eosinophil cationic protein is a diagnostic and predictive biomarker in young adults with inflammatory bowel disease. J Clin Med. 2019;8(12):2025.
- Trasolini R, Zhu K, Klemm N, et al. Fecal leukocyte esterase, an alternative biomarker to fecal calprotectin in inflammatory bowel disease: a Pilot series. Gastro Hep Adv. 2022;1(1):45–51.
- Shinzaki S, Matsuoka K, Iijima H, et al. Leucine-rich alpha-2 glycoprotein is a serum biomarker of mucosal healing in ulcerative colitis. J Crohns Colitis. 2017;11(1):84–91.
- Nakov R, Velikova T, Nakov V, et al. Trefoil factor 3 is highly predictive of complete mucosal healing independently and in combination with C-Reactive protein in patients with ulcerative colitis. J Gastrointest Liver Dis JGLD. 2019;28:169–174. doi: 10.15403/jgld-177
- Wakai M, Hayashi R, Tanaka S, et al. Serum amyloid a is a better predictive biomarker of mucosal healing than C-reactive protein in ulcerative colitis in clinical remission. BMC Gastroenterol. 2020;20(1):85.
- Jaenisch SE, Abbott CA, Gorrell MD, et al. Circulating Dipeptidyl peptidase activity is a potential biomarker for inflammatory bowel disease. Clin Transl Gastroenterol. 2022;13(1):e00452.
- Pinto-Lopes P, Afonso J, Pinto-Lopes R, et al. Serum Dipeptidyl peptidase 4: a predictor of disease activity and prognosis in inflammatory bowel disease. Inflamm Bowel Dis. 2020;26(11):1707–1719.
- Maeda K, Nakamura M, Yamamura T, et al. Gelsolin as a potential biomarker for endoscopic activity and mucosal healing in ulcerative colitis. Biomedicines. 2022;10(4):872.
- Shiraishi K, Furukawa S, Yagi S, et al. Serum globulin is associated with endoscopic findings and mucosal healing in Japanese patients with ulcerative colitis. Dig Dis Sci. 2022;67(1):233–240.
- Verstockt S, Verstockt B, Machiels K, et al. Oncostatin M is a biomarker of diagnosis, worse disease prognosis, and therapeutic nonresponse in inflammatory bowel disease. Inflamm Bowel Dis. 2021;27(10):1564–1575. doi: 10.1093/ibd/izab032
- Guo A, Ross C, Chande N, et al. High oncostatin M predicts lack of clinical remission for patients with inflammatory bowel disease on tumor necrosis factor α antagonists. Sci Rep. 2022;12(1):1185.
- West NR, Hegazy AN, Owens BMJ, et al. Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor–neutralizing therapy in patients with inflammatory bowel disease. Nat Med. 2017;23(5):579–589.
- Nishida Y, Hosomi S, Watanabe K, et al. Serum interleukin-6 level is associated with response to infliximab in ulcerative colitis. Scand J Gastroenterol. 2018;53(5):579–585.
- Bertani L, Rossari F, Barberio B, et al. Novel prognostic biomarkers of mucosal healing in ulcerative colitis patients treated with anti-TNF: neutrophil-to-lymphocyte ratio and platelet-to-lymphocyte ratio. Inflamm Bowel Dis. 2020;26(10):1579–1587.
- Mortensen JH, Godskesen LE, Jensen MD, et al. Fragments of citrullinated and MMP-degraded vimentin and MMP-degraded type III collagen are Novel serological biomarkers to differentiate Crohn’s disease from ulcerative colitis. J Crohns Colitis. 2015;9(10):863–872.
- Domislovic V, Høg Mortensen J, Lindholm M, et al. Inflammatory biomarkers of extracellular matrix remodeling and disease activity in Crohn’s disease and ulcerative colitis. J Clin Med. 2022;11(19):5907.
- Kalla R, Adams AT, Bergemalm D, et al. Serum proteomic profiling at diagnosis predicts clinical course, and need for intensification of treatment in inflammatory bowel disease. J Crohns Colitis. 2021;15(5):699–708.
- Sudhakar P, Salomon B, Verstockt B, et al. DOP79 biomarkers for IBD using OLINK proteomics inflammation panel: preliminary results from the COLLIBRI consortium. J Crohns Colitis. 2022;16(Supplement_1):i123–i124.
- Bergemalm D, Andersson E, Hultdin J, et al. Systemic Inflammation in Preclinical Ulcerative Colitis. Gastroenterology. 2021;161(5):1526–1539.e9.
- Torres J, Petralia F, Sato T, et al. Serum biomarkers identify patients who will develop inflammatory bowel diseases up to 5 years before diagnosis. Gastroenterology. 2020;159(1):96–104.
- Bourgonje AR, von Martels JZH, Gabriëls RY, et al. A combined set of four serum inflammatory biomarkers reliably predicts endoscopic disease activity in inflammatory bowel disease. Front Med. 2019;6:251. doi: 10.3389/fmed.2019.00251
- Baldan-Martin M, Azkargorta M, Iloro I, et al. P018 proteomic profile of serum and urine in newly diagnosed patients with inflammatory bowel disease: new approach for biomarker discovery. J Crohns Colitis. 2022;16(Supplement_1):i145.
- Andersson E, Bergemalm D, Kruse R, et al. Subphenotypes of inflammatory bowel disease are characterized by specific serum protein profiles. PLoS One. 2017;12(10):e0186142.
- Heier CR, Fiorillo AA, Chaisson E, et al. Identification of pathway-specific serum biomarkers of response to glucocorticoid and infliximab treatment in children with inflammatory bowel disease. Clin Transl Gastroenterol. 2016;7(9):e192.
- Adams A, Gupta V, Mohsen W, et al. Early management of acute severe UC in the biologics era: development and international validation of a prognostic clinical index to predict steroid response. Gut. 2023;72(3):433–442.
- Grant RK, Jones G-R, Plevris N, et al. The ACE (albumin, CRP and endoscopy) index in acute colitis: a simple clinical index on admission that predicts outcome in patients with acute ulcerative colitis. Inflamm Bowel Dis. 2021;27(4):451–457.
- Gibson DJ, Hartery K, Doherty J, et al. Crp/albumin ratio: an early predictor of steroid responsiveness in acute severe ulcerative colitis. J Clin Gastroenterol. 2018;52(6):e48–e52.
- Header DA, Aboelwafa RA, Elkeleny MR, et al. C-reactive protein/albumin ratio (CAR) as a marker for detecting acute severe ulcerative colitis in Egyptian patients. Rev Gastroenterol Mex Engl. 2022;87(4):447–454.
- Yagi S, Furukawa S, Shiraishi K, et al. The albumin to globulin ratio is associated with clinical outcome in Japanese patients with ulcerative colitis. Ann Coloproctology. 2023;39(2):155–163.
- Kalla R, Adams AT, Nowak JK, et al. Analysis of systemic epigenetic alterations in inflammatory bowel disease: defining geographical, genetic and immune-inflammatory influences on the circulating methylome. J Crohns Colitis. 2023;17(2):170–184.
- Gasparetto M, Payne F, Nayak K, et al. Transcription and DNA methylation patterns of blood-derived CD8+ T cells are associated with age and inflammatory bowel disease but do not predict prognosis. Gastroenterology. 2020;160(1):232–244.e7.
- Adams AT, Kennedy NA, Hansen R, et al. Two-stage genome-wide methylation profiling in childhood-onset Crohnʼs disease implicates epigenetic alterations at the VMP1/MIR21 and HLA loci. Inflamm Bowel Dis. 2014;20(10):1784–1793.
- Kalla R, Adams AT, Satsangi J. Blood-based DNA methylation in Crohn’s disease and severity of intestinal inflammation. Transl Gastroenterol Hepatol. 2019;4:76. doi: 10.21037/tgh.2019.10.03
- Ventham NT, Kennedy NA, Adams AT, et al. Integrative epigenome-wide analysis demonstrates that DNA methylation may mediate genetic risk in inflammatory bowel disease. Nat Commun. 2016;7(1):13507.
- Joustra V, Hageman IL, Satsangi J, et al. Systematic review and meta-analysis of peripheral blood DNA methylation studies in inflammatory bowel disease. J Crohns Colitis. 2023;17(2):185–198.
- Venkateswaran S, Somineni HK, Matthews JD, et al. Longitudinal DNA methylation profiling of the rectal mucosa identifies cell-specific signatures of disease status, severity and clinical outcomes in ulcerative colitis cell-specific DNA methylation signatures of UC. Clin Epigenetics. 2023;15(1):50.
- Burakoff R, Chao S, Perencevich M, et al. Blood-based biomarkers can differentiate ulcerative colitis from crohnʼs disease and noninflammatory diarrhea. Inflamm Bowel Dis. 2011;17(8):1719–1725.
- Biasci D, Lee JC, Noor NM, et al. A blood-based prognostic biomarker in IBD. Gut. 2019;68(8):1386–1395.
- Nowak JK, Adams AT, Kalla R, et al. Characterization of the circulating transcriptomic landscape in inflammatory bowel disease provides evidence for dysregulation of multiple transcription factors including NFE2, SPI1, CEBPB, and IRF2. J Crohns Colitis. 2022;16(8):1255–1268.
- Kalla R, Adams AT, Ventham NT, et al. Whole blood profiling of T-cell derived miRNA allows the development of prognostic models in inflammatory bowel disease. J Crohns Colitis. 2020;14(12):1724–1733.
- Yarani R, Shojaeian A, Palasca O, et al. Differentially expressed miRnas in ulcerative colitis and Crohn’s disease. Front Immunol. 2022;13:865777. doi: 10.3389/fimmu.2022.865777
- Zhou J, Liu J, Gao Y, et al. miRNA-Based potential biomarkers and new molecular insights in ulcerative colitis. Front Pharmacol. 2021 [cited 2023 Aug 13]; 12. Internet: 10.3389/fphar.2021.707776
- Vatn SS, Lindstrøm JC, Moen AEF, et al. Mucosal gene transcript signatures in treatment naïve inflammatory bowel disease: a comparative analysis of disease to symptomatic and healthy controls in the European IBD-Character cohort. Clin Exp Gastroenterol. 2022;15:5–25. doi: 10.2147/CEG.S343468
- Ostrowski J, Dabrowska M, Lazowska I, et al. Redefining the practical utility of blood transcriptome biomarkers in inflammatory bowel diseases. J Crohns Colitis. 2019;13(5):626–633.
- Wu Z, Bi Y. Potential role of MALT1 as a candidate biomarker of disease surveillance and treatment response prediction in inflammatory bowel disease patients. J Clin Lab Anal. 2022;36(2):e24130. doi: 10.1002/jcla.24130
- Mo A, Nagpal S, Gettler K, et al. Stratification of risk of progression to colectomy in ulcerative colitis via measured and predicted gene expression. Am J Hum Genet. 2021;108(9):1765–1779.
- Clerc F, Novokmet M, Dotz V, et al. Plasma N-Glycan signatures are associated with features of inflammatory bowel diseases. Gastroenterology. 2018;155(3):829–843.
- Shubhakar A, Jansen B, Adams A, et al. DOP10 serum N-glycomic biomarkers predict treatment escalation in inflammatory bowel disease. J Crohns Colitis. 2019;13(Supplement_1):S032–S033.
- Shubhakar A, Jansen BC, Adams AT, et al. Serum N-Glycomic biomarkers predict treatment escalation in inflammatory bowel disease. J Crohns Colitis. 2023;17(6):919–932.
- Pereira MS, Maia L, Azevedo LF, et al. A [Glyco]biomarker that predicts failure to Standard therapy in ulcerative colitis patients. J Crohns Colitis. 2019;13(1):39–49.
- Probert F, Walsh A, Jagielowicz M, et al. Plasma nuclear magnetic resonance Metabolomics discriminates between high and low endoscopic activity and predicts progression in a prospective cohort of patients with ulcerative colitis. J Crohns Colitis. 2018;12(11):1326–1337.
- Ghimire H, Viennois E, Hu X, et al. Infrared spectrometric biomarkers for ulcerative colitis screening using human serum samples. J Biophoto. 2022;15(6):e202100307.
- Berry N, Sinha SK, Bhattacharya A, et al. Role of positron emission tomography in assessing disease activity in ulcerative colitis: comparison with biomarkers. Dig Dis Sci. 2018;63(6):1541–1550.
- Borren NZ, Plichta D, Joshi AD, et al. Multi-“-omics” profiling in patients with quiescent inflammatory bowel disease identifies biomarkers predicting relapse. Inflamm Bowel Dis. 2020;26(10):1524–1532.
- Integrated bile acid profile analysis for the non-invasive staging diagnosis of ulcerative colitis. medRxiv [Internet]. [cited 2022 Nov 8]. Available from: https://www.medrxiv.org/content/10.1101/2022.03.14.22272391v1
- Sun Q, Tang Y, Dai L, et al. Serum bile acid metabolites predict the therapeutic effect of Mesalazine in patients with ulcerative colitis. J Proteome Res. 2023;22(4):1287–1297.
- Jagt JZ, Struys EA, Ayada I, et al. Fecal amino acid analysis in newly diagnosed pediatric inflammatory bowel disease: a multicenter case-control study. Inflamm Bowel Dis. 2022;28(5):755–763.
- Zhou G, Liu H, Wei P, et al. Amino acids-targeted metabolomics reveals novel diagnostic biomarkers for ulcerative colitis and Crohn’s disease. Amino Acids. 2023;55(3):349–358.
- Liu Z, Tang H, Liang H, et al. Dyslipidaemia is associated with severe disease activity and poor prognosis in ulcerative colitis: a retrospective cohort study in China. Nutrients. 2022;14(15):3040.
- Manfredi M, Conte E, Barberis E, et al. Integrated serum proteins and fatty acids analysis for putative biomarker discovery in inflammatory bowel disease. J Proteomics. 2019;195:138–149. doi: 10.1016/j.jprot.2018.10.017
- Galipeau HJ, Caminero A, Turpin W, et al. Novel fecal biomarkers that precede clinical diagnosis of ulcerative colitis. Gastroenterology. 2021;160(5):1532–1545.
- Iwasawa K, Suda W, Tsunoda T, et al. Dysbiosis of the salivary microbiota in pediatric-onset primary sclerosing cholangitis and its potential as a biomarker. Sci Rep. 2018;8(1):5480.
- Olbjørn C, Cvancarova Småstuen M, Thiis-Evensen E, et al. Fecal microbiota profiles in treatment-naïve pediatric inflammatory bowel disease – associations with disease phenotype, treatment, and outcome. Clin Exp Gastroenterol. 2019;12:37–49. doi: 10.2147/CEG.S186235
- Olbjørn C, Småstuen MC, Moen AEF. Targeted analysis of the Gut microbiome for diagnosis, prognosis and treatment individualization in pediatric inflammatory bowel disease. Microorganisms. 2022;10(7):1273. doi: 10.3390/microorganisms10071273
- Vatn S, Carstens A, Kristoffersen AB, et al. Faecal microbiota signatures of IBD and their relation to diagnosis, disease phenotype, inflammation, treatment escalation and anti-TNF response in a European multicentre study (IBD-Character). Scand J Gastroenterol. 2020;55(10):1146–1156.
- Liu J, Fang H, Hong N, et al. Gut microbiome and metabonomic profile predict early remission to anti-integrin therapy in patients with moderate to severe ulcerative colitis. Microbiol Spectr. 2023;11(3):e0145723.
- Rees NP, Shaheen W, Quince C, et al. Systematic review of donor and recipient predictive biomarkers of response to faecal microbiota transplantation in patients with ulcerative colitis. EBioMedicine. 2022;81:104088. doi: 10.1016/j.ebiom.2022.104088
- van Thiel IAM, Rahman S, Hakvoort TBM, et al. Fecal Filobasidium is associated with clinical remission and endoscopic response following fecal microbiota transplantation in mild-to-moderate ulcerative colitis. Microorganisms. 2022;10(4):737.
- Nishida Y, Hosomi S, Yamagami H, et al. Novel prognostic biomarkers of pouchitis after ileal pouch-anal anastomosis for ulcerative colitis: neutrophil-to-lymphocyte ratio. PLoS One. 2020;15(10):e0241322.
- Okba AM, Amin MM, Abdelmoaty AS, et al. Neutrophil/Lymphocyte ratio and lymphocyte/monocyte ratio in ulcerative colitis as non-invasive biomarkers of disease activity and severity. Autoimmun Highlights. 2019;10(1):4.
- Kurimoto N, Nishida Y, Hosomi S, et al. Neutrophil-to-lymphocyte ratio may predict clinical relapse in ulcerative colitis patients with mucosal healing. PLoS One. 2023;18(1):e0280252.
- Anderson A, Cherfane C, Click B, et al. Monocytosis is a biomarker of severity in inflammatory bowel disease: analysis of a 6-year prospective natural history registry. Inflamm Bowel Dis. 2022;28(1):70–78.
- Gettler K, Levantovsky R, Moscati A, et al. Common and rare variant prediction and penetrance of IBD in a large, multi-ethnic Health System-Based Biobank Cohort. Gastroenterol. 2021;160(5):1546–1557.
- Kikut J, Mokrzycka M, Drozd A, et al. Involvement of proinflammatory arachidonic acid (ARA) derivatives in Crohn’s disease (CD) and ulcerative colitis (UC). J Clin Med. 2022;11(7):1861.
- Debourdeau E, Chamard C, Carriere I, et al. Retinal microcirculation changes in Crohn’s disease patients under biologics, a potential biomarker of severity: a Pilot study. J Pers Med. 2022;12(2):230.
- Zittan E, Steinhart AH, Aran H, et al. The Toronto IBD Global endoscopic Reporting [TIGER] score: a single, easy to use endoscopic score for both Crohn’s disease and ulcerative colitis patients. J Crohns Colitis. 2022;16(4):544–553.
- Rowan CR, Cullen G, Mulcahy HE, et al. DUBLIN [degree of ulcerative colitis burden of Luminal inflammation] score, a simple method to quantify inflammatory burden in ulcerative colitis. J Crohns Colitis. 2019;13(11):1365–1371.
- Gordon IO, Agrawal N, Willis E, et al. Fibrosis in ulcerative colitis is directly linked to severity and chronicity of mucosal inflammation. Aliment Pharmacol Ther. 2018;47(7):922–939.
- Mortensen JH, Lindholm M, Langholm LL, et al. The intestinal tissue homeostasis – the role of extracellular matrix remodeling in inflammatory bowel disease. Expert Rev Gastroenterol Hepatol. 2019;13(10):977–993.
- Boyapati RK, Dorward DA, Tamborska A, et al. Mitochondrial DNA is a pro-inflammatory Damage-associated molecular Pattern Released During active IBD. Inflamm Bowel Dis. 2018;24(10):2113–2122.
- Mitochondrial DAMPs as mechanistic biomarkers of mucosal inflammation in Crohn’s disease: study protocol for prospective longitudinal cohort study in Scotland. medRxiv [Internet]. [cited 2022 Nov 8]. Available from: https://www.medrxiv.org/content/10.1101/2022.03.21.22270313v1
- Argmann C, Hou R, Ungaro RC, et al. Biopsy and blood-based molecular biomarker of inflammation in IBD. Gut [Internet]. 2022 [cited 2023 Jan 17]. Available from: https://gut.bmj.com/content/early/2022/09/14/gutjnl-2021-326451
- Khanna R, Narula N, Feagan BG. The role of biomarkers in clinical trials of inflammatory bowel disease. Inflamm Bowel Dis. 2018;24(8):1619–1623. doi: 10.1093/ibd/izy195
- Gabriëls RY, Bourgonje AR, von Martels J, et al. P577 Mucosal eosinophil abundance in non-inflamed colonic tissue predict response to vedolizumab induction therapy in inflammatory bowel disease. J Crohns Colitis. 2022;16(Supplement_1):i517–i518.
- Roosenboom B, Wahab PJ, Smids C, et al. Mucosal α4β7+ Lymphocytes and MAdCAM+ Venules Predict Response to Vedolizumab in Ulcerative Colitis. Inflamm Bowel Dis. 2023;zad123. doi: 10.1093/ibd/izad123
- Telesco SE, Brodmerkel C, Zhang H, et al. Gene expression signature for prediction of Golimumab response in a phase 2a open-label trial of patients with ulcerative colitis. Gastroenterology. 2018;155(4):1008–1011.e8.
- Scheer J. Gould and Goldman: deadly deceit--a defense. Health Phys. 1991;61(2):279–280.
