ABSTRACT
Introduction
Diagnostics are an essential, undervalued part of the health-care system. For many diseases, molecular diagnostics are the gold standard, but are not easy to implement in Low- and Middle-Income Countries (LMIC). Sample-to-result (S2R) platforms combining all procedures in a closed system could offer a solution. In this paper, we investigated their suitability for implementation in LMIC.
Areas covered
A scorecard was used to evaluate different platforms on a range of parameters. Most platforms scored fairly on the platform itself, ease-of-use and test consumables; however, shortcomings were identified in cost, distribution and test panels tailored to LMIC needs. The diagnostic coverage for common infectious diseases was found to have a wider coverage in high-income countries (HIC) than LMIC. A literature study showed that in LMIC, these platforms are mainly used as diagnostic tools or evaluation of diagnostic performance, with a minority assessing the operational characteristics or the clinical utility. In this narrative review, we identified various points for adaptation of S2R platforms to LMIC conditions.
Expert opinion
For S2R platforms to be suitable for implementation in LMIC some modifications by the manufacturers could be considered. Furthermore, strengthening health systems and digitalization are vital; as are smaller, cheaper, faster, and sustainable technologies.
1. Introduction
Access to appropriate diagnostic tools is an essential component in the evaluation and improvement of global health [Citation1]. Although laboratory diagnostics are crucial to health-care systems, the attention and priority given by funders and policymakers is low in comparison to new therapeutics, vaccines, or strategies for prevention [Citation1,Citation2]. The World Health Organization (WHO) has had its essential medicines list since 1977; an equivalent essential diagnostics list was only published in 2018 and no plans for diagnostics or their financing have been found in the available implementation plans for universal health coverage [Citation2].
The main limitation to access to diagnostic tools in many countries is the inequitable and poor distribution. In 2015, 47% of the world’s population had little to no access to basic diagnostics, while in low and middle-income countries (LMIC) this was as high as 81% at the primary care level [Citation2]. This limited access to laboratory diagnostic tests is a serious obstacle to equity in health services [Citation3,Citation4] and detrimental to optimal health-care provision in low-resource settings (LRS) [Citation5,Citation6].
For communicable diseases, lab-based diagnostics are vitally important not only for case management on an individual patient level but also for disease control on a public health level [Citation1,Citation2]. A diagnostic decision based on clinical symptoms is often the only option available for health-care workers in LRS, but has been shown to be inaccurate and hamper effective disease control measures and case management [Citation7,Citation8]. Accurate and rapid diagnosis improves patient care [Citation9,Citation10] and guide to a more specific treatment leading to a reduction in morbidity and disease transmission [Citation11,Citation12]. Furthermore, it could also advance epidemiological surveillance, timely detection of outbreaks, and implementation of control measures [Citation6,Citation13], such as, vector control or quarantines [Citation14].
The 2013–2016 West Africa Ebola outbreak and the more recent COVID-19 pandemic have shown impressively the importance of a good diagnostic infrastructure in terms of limiting pathogen transmission. With good and rapid diagnostics in place, outbreaks can be detected timely and controlled more quickly. This will lead to fewer infections, lowered costs for the health system and a faster return to usual economic activity, hence less economic losses [Citation2,Citation15,Citation16].
With their high sensitivity and specificity, molecular diagnostics are the preferred diagnostic tool for a lot of pathogens causing infectious diseases, especially acute infections [Citation17–19]. However, in many health-care facilities in low-resource settings, they are not available. The lack of molecular tests for clinicians and patients in health-care facilities has several reasons. The necessary equipment is costly and needs maintenance, most often by trained technicians [Citation5,Citation6]. Extraction of nucleic acids and real-time PCR require a high standard of laboratory infrastructure, such as highly trained staff and separated work spaces for different steps of the process to avoid contamination [Citation10,Citation20,Citation21]. When PCR tests are not available at the point of care, samples have to be sent to reference laboratories resulting in long turn-around times (TAT). Not only does this discourage patients from going for testing but it often makes them leave without a lab-confirmed diagnosis and only syndromic treatment [Citation4,Citation22]. Despite this, even in low-resource settings the implementation of molecular diagnostics is possible, as the COVID-19 pandemic has shown. In the WHO African Region, during the pandemic the number of laboratories able to diagnose SARS-CoV-2 with molecular tests went from 2 in February 2020 to 750 in November 2020 [Citation23]. Some examples for the increase of laboratories from LMIC include Nigeria [Citation24], Benin [Citation25], India [Citation26] and Bangladesh [Citation27].
Sample-to-result (S2R) platforms for molecular diagnosis can mitigate some of the problems encountered with standard real-time PCR for low-resource settings. A S2R platform is defined as a system where nucleic acid extraction, nucleic acid amplification, and analysis are performed in a closed system. All necessary reagents and controls are incorporated in disposable cartridges or cassettes. Hands-on time for sample preparation is minimal, and the closed system decreases the risk of contamination. Lab technicians without training in molecular diagnostics can perform these tests reliably [Citation21,Citation22,Citation28]. There is no necessity to place them in a laboratory specialized for molecular diagnostics [Citation21], but they do need a stable electricity supply during their run and a temperature-controlled environment [Citation29,Citation30].
As with traditional real-time PCR, one limiting factor LMIC is the cost, as the disposable cartridges or cassettes are often more expensive than standard PCR and extraction [Citation31]. However, it is important to consider cost savings when automating the process as it allows for reduced human resource (HR) expenses, freeing up the lab technician to focus on other tasks. With higher HR costs in High-Income Countries (HIC) settings this HR cost aspect is relevant primarily for HIC settings and less so for LMIC, the major advantages of S2R platforms for LMIC are that the needs for the level of training for operators is significantly lower and the quality of the test result can be better assured.
Another factor is reduced health-care costs, resulting from a test with a quick TAT, and high sensitivity and specificity. The Xpert MTB/RIF assay has proven this to be true for tuberculosis (TB) by significantly reducing all costs occurring with a wrong or delayed diagnosis [Citation7,Citation21,Citation22]. With a WHO recommendation and a discounted price for 116 countries negotiated by FIND, the GeneXpert® platform has already shown that its implementation is possible in low-resource settings and its utility has been proven for diagnosis of tuberculosis and HIV. In LMIC it is now the most common system for molecular testing [Citation22]. However, its implementation was more successful in UMIC than in LMIC due to several implementation challenges [Citation32].
Apart from the GeneXpert for TB and HIV, which are vertically funded programs, implementation of these machines in LMIC has been slow and their impact poorly evaluated. Contrary to other point-of-care tests compliant with the ASSURED criteria (affordable, sensitive, specific, user friendly, rapid and robust, equipment-free, and deliverable [Citation11]), there is a lack of a definition for the suitability of S2R platforms for LMIC. In this narrative review, we assess several of the most common S2R molecular diagnostic platforms and evaluate their suitability for implementation in LMIC settings to diagnose infectious pathogens of concern to public health. The minimal requirements for an S2R platform to be used in an LMIC setting include the ease of use, a fast turn-around time, compatibility with different sample types, and offering a wide range of test panels that are relevant for the different regions. Furthermore, distribution and technical servicing should be easily available in the country with a reasonable price range for the equipment and consumables. The S2R pathogen panels should be validated for the pathogens in the regions and evaluated on their clinical utility and public health impact. Integration of the platform in the health-care system with treatment or referral options is also very important. Several parameters were taken into account. Firstly, the platforms were compared with a scorecard across several parameters for their operational characteristics. Secondly, to assess if their diagnostic coverage is adequate for LMIC, the pathogen panels available on each platform were correlated with pathogens of importance in different income and geographical regions. Thirdly, a literature search on the existing use of these platforms in LMIC was performed to investigate which machines are used in which countries and what kind of studies have been published.
2. Sample-to-result (S2R)
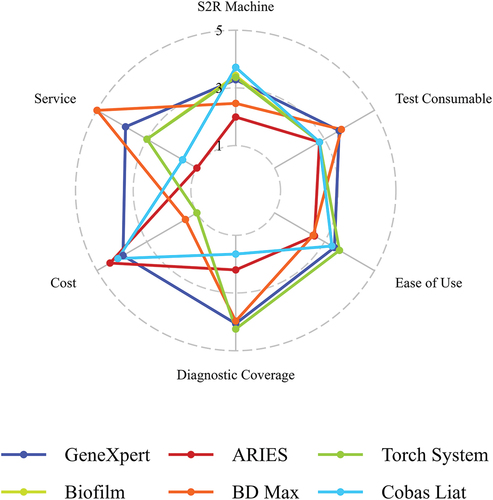
The criteria for selecting and including S2R platforms into our review are: (1) all steps for extraction and real-time PCR (with hydrolysis or melt-point analysis) are included in one cartridge with no separate extraction needed, and (2) their commercial availability with several different pathogen panels. An exhaustive list of machines on the market fit these criteria; however, in this review we focus on the most common platforms, as defined by the number of publications and available test menu, i.e. GeneXpert XVI (Cepheid), ARIES® M12 (Luminex/Diasorin), BioFire® (BioMerieux), BD MAX™ (BD), and Cobas® Liat® (Roche). More details can be found in and supplementary data.
Table 1. Selection criteria of Sample-To-Result (S2R) platforms.
2.1. S2R platform assessment with a scorecard
2.1.1. Selection criteria for S2R platforms
As selection criteria, we defined at least 20 publications, the available test menu (extending beyond respiratory pathogens), instrument size and weight (to assess ease of transport). This review limits itself to test panels for infectious diseases, excluding other commonly available tests for cancers. As such, S2R platforms only offering respiratory panels (ID Now, Abbott; VERSANT® kPCR Molecular System, Siemens Healthineers) or not offering infectious disease assays (Idylla®, Biocartis) were excluded. Furthermore, large and heavy platforms (Cobas 800, GeneXpert Infinity), that cannot be placed on a counter were also not considered. S2R platforms using other amplification technology such as LAMP (Simprova, Eiken Chemical) or helicase dependent (Quidel Solana) were also excluded.
The GeneXpert platform was included in this review due to its worldwide distribution, its extensive infectious disease test menu, and its WHO recommendations for use in TB and HIV programs. The BioFire platforms (Torch system and BioFilm v2.0) were included because BioMérieux’s offering a broad infectious disease test menu and presence in some LMIC with a high number of publications. BD Max has an extensive infectious disease test panel, a high number of publications and provides an ‘open’ system (allowing the user to use the cartridges with any PCR). Although the Cobas Liat does not have an extensive pathogen test panel (only one pathogen other than respiratory), it was included to represent a POC version of a light-weight small footprint S2R with very fast TAT. Furthermore, Roche Diagnostics is a known diagnostics company whose widely used Cobas 800 was excluded due to its size and there are further tests in the pipeline (Pertussis and HIV) for the Cobas Liat [Citation33]. ARIES (Luminex/DiaSorin) was included because it offers an open cartridge system and allows to implement in-house assays on a S2R platform.
2.1.2. Scoring of criteria
To evaluate the operational characteristics of the S2R platforms, a scorecard was adapted from a previously published scorecard for POC assays [Citation34], and each of the products was assessed according to predetermined specifications, encompassing aspects such as the weight, overall dimensions, test throughput, test menu, running time, service, and maintenance accessibility, user friendliness, and pricing.
Six categories were chosen with 32 criteria in total.
S2R platform: specifications of the platforms, such as dimensions, running time, installation etc.
Test consumables: appropriateness of storage and waste disposal of consumables.
Ease of use: level of operator skills and amount of hands-on time and extra equipment needed.
Diagnostic coverage: Adequacy of offered pathogen panels for LMIC needs and number of pathogens offered in general.
Cost: cost of the S2R platform and its consumables
Distribution and service: availability of distribution and service available in mostly LMICs
The S2R platforms were scored on all parameters from 1 to 5 (with 5 being the best) to the parameters defined important for LMIC. The final score was calculated from the individual weighted scores. The input for all the different parameters was researched in publicly available websites and company documents. For continuous variables such as weight, dimensions, costs, or number of offered pathogens the median and the first and third quartile were calculated and used for scoring. For dichotomous variables (Yes/No) such as quantification possible or is it an open system, 5 points were assigned for yes and 1 for no. For categorical variables scores were assigned depending on their specifications. For example, for the additional equipment, 5 points were assigned if no additional equipment was needed, 4 if only easily available equipment was needed such as pipettes and vortex, 3 points were assigned if additionally to pipettes and vortex, special consumables were needed (special swabs for sample transfer), 3 points if a centrifuge was needed and 1 point if a heater was needed. Different weights were assigned to the parameters depending on their importance. A detailed table with all the categories and parameters for each S2R platform with references can be found in supplementary data .
The scorecard design with all the categories, their criteria, scoring, and weights is presented in . There are several limitations to this scorecard rating. Only five of the most common S2R systems were taken into account and in no intention this review is meant to provide a complete and exhaustive overview of platforms on the market. For the scorecard evaluation only publicly available data was used, but some of this data was not easily available and might not be reflecting the real data or vary depending on the location, which includes data on costs, shelf life, or availability of pathogen panels. The ratings for the scorecard were done based on literature and the publicly available documents and not on the practical experience of the users. The scorecard also covered some criteria less in-depth, such as for ‘cost.’ The costs related to maintenance or purchase of control kits was not taken into account, as this information is not easily retrievable from online sources and price setting can differ significantly depending on the region, customer–company relationship, and marketing strategy. The S2R platforms were also compared over a range of offered assays with different running times, sample types, and specimen preparation procedures.
Table 2. Categories and criteria of the scorecard with their weights. Open system is referring to cartridges where the user is free to use any PCR. In different versions available, stackable refers to the fact that different modules can be stacked on top of each other to save space.
2.1.3. S2R platform
With 5 points being the maximum score in the category ‘S2R platform criteria,’ the BioFire (3.43), the GeneXpert (3.20), and Cobas Liat (3.71) platforms scored similarly, with BD Max (2.46) and ARIES (1.99) on the lower end. The S2R platforms varied substantially in weight and overall dimensions with Cobas Liat being the smallest, but also with lowest throughput and BD Max being the biggest but with highest test throughput. In an LMIC, a smaller machine is advantageous since it occupies less space, which is often limited in a laboratory. However, for achieving a quick TAT for results, a high throughput is just as crucial as the running time ().
In the Cobas Liat platform, only one sample at a time can be processed, but the platform’s efficient 20-min running time compensates for this limitation. The mean running time of different assays for GeneXpert and BioFire systems was around 1 h, for ARIES 2 h and for BD Max almost 2.5 h. Cepheid’s provision of an early assay termination feature for certain tests, which halts the run as soon as the pathogen is detected, is a great way of reducing the running time. The ability to run individual tests on these platforms offers significant flexibility in processing samples for various tests as soon as they arrive in the lab, which is where GeneXpert, BioFire and partly the ARIES score high. The ARIES system has two modules with six slots each, so two different tests can be run for a maximum of six samples. Short running times or individual runs are advantageous, as single samples from patients can be run at once for a short TAT and rapid communication of results.
Flexibility in S2R platform versions is essential for LMIC, as health-care providers with a lower volume of tests to process can avoid the expense of acquiring high-throughput machines. The GeneXpert offers a range of versions with varying module capacities, such as 2, 4, and 16 modules. Similarly, the BioFire platforms, including the Torch system with a maximum capacity of 10 modules and the BioFilm versions with 8 modules, offer the advantage of expanding the number of modules as needed by being stacked upon each other, thereby accommodating changing requirements.
For most diseases, a qualitative test is sufficient since determining the presence or absence of a disease is the primary concern for effective case management. For some diseases, however, such as HIV, HBV, and HCV the viral load is a crucial component for disease management. GeneXpert offers three assays that provide quantitative PCR (HIV, HBV, HCV) and BioFire one. On the BD Max, quantification is technically possible according to the instrument specifications, but no quantitative tests were found that are commercially available.
Given that all of the companies are based in HIC and may not prioritize developing tests for LMIC [Citation1], an ‘open’ system would be a game-changer. This would empower users to transfer in-house developed PCRs to an S2R platform or develop and validate new assay panels in accordance with the specific disease requirements in their setting [Citation35]. The only two platforms offering an ‘open’ system are the ARIES and BD Max. In the open system from ARIES assays for BK Virus [Citation36], Pneumocystis jirovecii [Citation37], MERS-CoV [Citation38], and Ehrlicha spp./Anaplasma phagocytophilium [Citation39] were developed and validated. In our lab, we developed a multiplex-PCR for arboviruses (ZIKV, DENV, CHIKV, YFV) on the ARIES platform (unpublished data). On the BD Max open system the company Certest offers tests for Pneumocystis jirovecii, Bordetella, vancomycin resistance, and different SARS-CoV-2 variants, additionally they provide a RUO assay for monkeypox [Citation40].
Since distribution and service is often compromised or not available in LMIC, an ideal S2R platform for LMIC would not need a company licensed service worker for installation and maintenance. However, most machines have to be installed and maintained by a company licensed service worker, with the exception of Cobas Liat. The company’s website specifies that GeneXpert needs to be maintained by a licensed service worker, but there are instructions found online for installation and maintenance with the calibration kits by the end-user. The Cobas Liat is also the only machine that does not need a preventative maintenance; however, the BioFire machines are maintained on an as-needed basis and the GeneXpert after 2,000 runs or annually, which is similar for the ARIES system.
2.1.4. Test consumables
In terms of test consumables BD Max (3.68) and GeneXpert (3.59) score best, whereas ARIES, BioFire systems and Cobas Liat have a lower score of 2.80. Most consumables can be stored at room temperature, only Cobas Liat consumables require a 2–8°C cold chain, along with the Xpert Norovirus from GeneXpert. It is worth noting that the maximum storage temperature was 25–30°C, and in tropical regions, exposure to higher temperature could be a concern. There is no data in the public domain on the evaluation of stability of cartridge reagents above 30°C. BD Max, GeneXpert and Cobas Liat offer tests with a higher shelf life of around 18 months on average, compared to approximately 12 months for ARIES and BioFire.
Another crucial but often overlooked aspect of test consumables is their proper disposal. Apart from the challenges related to disposing of plastics and biohazardous waste, the cartridges or cassettes commonly contain Guanidine-Thiocyanate (GTC), which can be very toxic to humans and the environment if not disposed correctly through incineration at high temperatures [Citation41–43]. The only indication for waste disposal found in the instructions was to treat the cassettes/cartridges as biohazardous and to dispose of them in accordance with the regulations of their respective countries. However, all but BD Max cassettes/cartridges contained GTC. BD Max uses a different chemical (4-(1,1,3,3-tetramethylbutyl)phenol, ethoxylated (4-tert-OPnEO, ‘OPnEO’)), which also requires incineration [Citation44]. Information regarding the proper disposal of waste generated by these platforms was easiest to retrieve for GeneXpert. NGOs or organizations working with GeneXpert have online resources providing guidance on waste disposal [Citation45,Citation46], but it was extremely difficult to find information online for the other S2R platforms.
Precise figures regarding the quantity of waste generated by S2R platforms cannot be found, except for some examples. By August 2020, COVID-19 PCR tests had already generated 15,000 tons of plastic waste [Citation47], and for the year 2020 the HIV VL testing produced 924,000 liters of chemical waste and 2.1 million kg of plastic waste [Citation43]. This might not matter too much in high-income settings, where strict regulations and appropriate facilities for waste disposal are available, but the situation is very different in LMIC. The push for decentralized testing often disregards the lack of waste disposal regulations and facilities in many countries [Citation43]. In a survey done in 11 African countries regarding HIV-VL use, Odhiambo et al. found that 9 out the 11 did not have guidelines or regulations for waste disposal [Citation43], and according to a Global Fund survey only 32% of health-care facilities had a formal waste management system, whereas 58% resorted to open burning or dumping [Citation42,Citation45,Citation48]. Chemical waste from Cepheid cartridges has been found to be poured in the drain, despite the availability of incineration options [Citation43]. Even if in vertical programs incinerators are put in place, as was the case during the West Africa Ebola outbreak, a sustainable plan for their continued use is often absent and many were non-operational a few years later. There is a clear lack of infrastructure, expertise but also regulatory frameworks and policies and their enforcement. While commendable efforts are being made to improve diagnostic access in LMICs, waste management is frequently overlooked or insufficiently addressed [Citation42]. This critical aspect is notably absent from the recent Lancet Commission on diagnostics [Citation2]. Although the problems are known to the global health advocacy community, while lack of infrastructure, such as human resources or equipment for the laboratory is taken into account, waste disposal often is not [Citation42]. The sole responsibility of waste management falls on the end-user in health-care facilities or countries; however, manufacturers should bear responsibility to guide end users and customers to proper waste disposal of their test kits [Citation43,Citation45]. As the use of S2R platforms with single-use consumables increases, the challenges associated with their proper disposal will only intensify. Nonetheless, for the mitigation of the impact of extensive health interventions on human well-being and the prevention of environmental consequences, sustainable healthcare waste management is indispensable [Citation42].
2.1.5. Ease of use
For ease of use the BioFire systems (3.60), GeneXpert (3.40) and Cobas Liat (3.30) scored the highest, with ARIES (2.60) and BD Max (2.55) a bit lower. The ease of use was similar over all S2R platforms as the required skills are not very advanced and only a few steps were necessary to prepare the various sample types. For different sample types different preparational steps were needed except for the BioFire systems. Most assays only required a vortex and pipette (BioFire, Cobas Liat) or additional swabs/loops for sample transfer (GeneXpert and BD Max) for some sample types. Only the ARIES had a centrifugation step for stool samples. With incubation time for samples such as sputum, hands on time increased to 20–40 min, while for other sample types it was only 2–5 min. The ease of use and short hands-on time are one of the biggest advantages of S2R systems, where all platforms performed similarly. Since the S2R platforms are easy to use, not a lot of training for the end-user is needed, which facilitates use and human resources, as end-users do not need to have specific expertise, other than basic laboratory skills [Citation30]. With lab technicians in short supply in LMIC and a high turnover rate this is a major advantage of the S2R platforms [Citation49]. The risk of contamination is reduced and the exposure to potentially biohazardous samples is significantly decreased. The short hands-on time also allows the laboratory technician to perform other tasks, while the S2R platforms are running which can contribute to the cost-effectiveness, as human resources can be used optimally [Citation21].
2.1.6. Diagnostic coverage/test menu
A big difference was seen in the scores for diagnostic coverage with BioMerieux offering the most diverse test menu with 4.25 points, followed by GeneXpert with 4.11 points and BD Max with 4.05 points. ARIES and Cobas Liat scored 2.25 points and 1.70 points, respectively, for their test menus. This is reflective of the number of panels and pathogens offered by the companies as this was highest for BioMerieux (132 pathogens in 14 panels), BD (35 pathogens in 26 panels), and Cepheid (25 pathogens in 35 panels). Luminex/Diasorin only had 13 pathogens in 9 panels, and Roche Liat only had 6 pathogens in 5 panels. The number of offered pathogens is also indicative of the high coverage of pathogens of medical importance in LMIC, which will be discussed further below, where BioFire and BD received highest rankings. Cepheid also scores high as some of their tests received WHO prequalification, whereas the others have FDA approval and/or CE marking. WHO prequalification is used in LMIC to assure the quality and safety of drugs, medical devices and diagnostics as relying only on the FDA or CE marking might not be sufficient for LMIC. These regulatory approvals are for the US-American and European market and diagnostic needs might be different in other regions of the world [Citation2].
2.1.7. Cost
In our cost analysis, we factored in the expenses related to equipment and consumables, excluding the costs for control or calibration kits. Finding the costs with publicly available information was not very straight-forward, so prices might vary. This lack of transparency when it comes to pricing can also be limiting when implementing S2R platforms, as it is not very easy to plan for a budget. For machines with several options of modules/size the biggest size was taken into account; for example, for the GeneXpert we considered the 16-slot configuration, even though smaller options (2 and 4 slots) were available at a lower cost. The systems of GeneXpert, ARIES and Cobas Liat are very similar in their price range of machines and consumables and therefore have similar scores (4.48, 5.00 and 4.48 respectively). BioMerieux’s machines offer stackable configurations with varying module numbers. For the Torch System and BioFilm v2.0 the only prices found online were for 2 modules [Citation50]. The BD Max had the second lowest score with 1.46 as it was the most expensive S2R, but in turn offers the highest throughput capacity with 24 samples at a time. When taking into account the throughput number by dividing the price by available sample slots, ARIES and GeneXpert were the cheapest and the BD Max only slightly more expensive. The BioFire systems were the most expensive hence received the lowest score (1.0). The price for consumables for the BioFire S2R and BD Max was substantially higher than the other S2R, but BioFire offered the highest number of pathogens covered in one assay (n = 34) compared to 4 or 5 pathogens with the other S2R platforms. Although out of the scope for this review, a comparison of the clinical performance of different S2R platforms in different settings and countries would be of interest. Affordability is a major problem in LMIC even if testing with the S2R platform is cost-effective comparing to the standard procedure [Citation51,Citation52]. As seen with the GeneXpert after the price was reduced from 16.68 US$ to 9.98US$ per cartridge for their TB test in 2012, uptake accelerated significantly [Citation30]. However, many LMIC TB programs cannot afford this price, as it is suggested that a TB test replacing microscopy should be 4-6US$ [Citation53]. Furthermore, a lot of hidden costs, such as adapting the laboratory infrastructure, purchase of energy back-up systems, security systems or custom clearances are often not calculated in the budgets or are not foreseeable [Citation54]. More studies investigating the cost-effectiveness or benefit-cost ratio in a range of different settings and diseases are needed, but difficult to do, especially for diseases where no treatment is available [Citation2]. Studies over public health impact, like via vector control, are even more complicated to perform.
Several mechanisms exist to lower the cost of equipment and consumables. Pooled purchasing as done by various NGOs with their vertical programs can lower prices significantly. This means purchasing over various countries and years [Citation2], which could be organized by some regional supra-national bodies such as Africa CDC [Citation55]. Another strategy could be long-term framework agreements, as shown in South Africa for HIV VL testing [Citation56,Citation57]. Giving a higher priority to diagnostics in policy making could also bring more financing to diagnostics and by adapting the WHO EDLs to a national EDL, tailored to the specific needs of the country, could help to prioritize this extra funding [Citation2]. Optimizing the allocation and utilization of resources could help governments to increase their spending in health, as 20–40% of resources are wasted [Citation57,Citation58]. All of these considerations are not exclusive to S2R platforms but valid for all kinds of diagnostics.
2.1.8. Distribution and service
For distribution and service BD (5.0), Cepheid (3.86) and BioMerieux (3.0) score the highest. BD has the highest score due to their widespread presence also in LMIC as a company (with the limitation that it is not certain if that extends to BD Max) and the remote service capability of the BD MAX. Cepheid has the highest number of countries in which they offer service and distribution of their products reflective of their involvement in TB and HIV testing in LMIC. BioMerieux has less presence in LMIC but offers software for the remote management and monitoring of the platform. Roche Liat and Luminex/Diasorin have the lowest scores with 1.57 and 1.0, respectively, as they have limited presence in LMIC. Roche offers software for the Cobas Liat for remote transfer of reports or downloading assays, but it is not clear if a remote service capability exists. Luminex/Diasorin has no remote service from the machine; the user has to download the error reports and make a manual case after e-mail contact. The provision of service in a limited number of countries for not only installation but also maintenance is one of the most significant challenges in LMIC. It might not be possible to obtain service or maintenance or even if it is available, it might not be in time or affordable [Citation2,Citation30]. This is a matter that often goes overlooked when S2R platforms are donated and can result in substantial issues as a portion of donated equipment may become inoperative and unusable [Citation59]. Consequently, this situation might necessitate the procurement of services from third-party providers, potentially impacting warranties [Citation2].
GeneXpert had the best overall score across categories with an average of 3.77 points (3.30–4.25). The BD Max (3.20 points (1.92–4.48), the BioFire system (BioFilm 3.01 points (1.90–4.12), Torch system 3.00 points (1.90–4.11)) and the Cobas Liat (2.93 points (1.78–4.07)) had similar scores. The lowest scores were achieved by the ARIES system with 2.61 points (1.28–3.94).
2.2. Do the offered test menus address diagnostic needs in LMIC?
To assess the adequacy of the offered test menu a comparative analysis of the available pathogen panels on the Global Burden of Disease (GBD) 2019 report [Citation60] was performed. This was done by comparing the diseases listed with the test menus of the companies. For the GBD 2019 report data was downloaded from vizhub [11 October 2023] [Citation60]. The following parameters were used for the download: GBD Estimate (Cause of death or injury/Etiology), Measure (Deaths, DALYs), Metric (Number, Percent, Rate), Age (all ages), Sex (both), and Year (2019). The scope of GBD estimate encompassed all level 4 causes, with only infectious diseases, across diverse global regions. These regions included World Bank categories such as High-Income Countries (HIC), Upper-Middle-Income Countries (UMIC), Lower-Middle-Income Countries (LMIC), Low-Income Countries (LIC). Another download was performed with geographical regions: Australasia, Central Asia, Central Europe, Central Sub-Saharan Africa, East Asia, Eastern Europe, Eastern Sub-Saharan Africa, High-Income Asia Pacific, High-Income North America, Latin America and Caribbean, North Africa, Middle East, Oceania, South Asia, South East Asia, Southern Latin America, Southern Sub-Saharan Africa, Western Europe, and Western Sub-Saharan Africa. Syndromic diseases with several potential causative pathogens, such as ‘upper respiratory infection’ were excluded. For the diseases on the GBD 2019 report, the gold standard diagnostic method was reviewed in the literature, as can be seen in supplementary table S2 diagnostics references. The focus was solely on diagnostic methods commonly employed for clinical diagnoses, thus excluding methods primarily designed for research or epidemiological studies. The income and geographical regions were sorted in descending order for the most frequent infectious diseases by Disability-adjusted life years (DALYs) by rate (DALYs per 100,000) and all diseases which are not commonly diagnosed by PCR were excluded. The results can be seen in supplementary table S3. The 10 most frequent diseases were then compared with test menus offered for each of the considered S2R platforms and presented visually through a bar graph plot. All data analysis and graphical representation were performed utilizing the R statistical software and QGIS, a geographic information system.
The total number of tests for the 10 most prevalent infectious diseases by DALYs offered over all S2R platforms was 28 for all HIC, 27 for UMIC, 20 for LMIC and 22 for LIC (out of 50). A detailed table with all diseases per income group and region can be seen in supplementary table S3. As seen in , the region’s best served by companies with the total number of tests offered by all companies for the 10 most prevalent diseases were Australasia, Central Europe, Northern America and Southern Latin America with over 28 tests in total (out of 50). These are also regions with mostly HIC. Central Asia, East Asia, High-Income Asia Pacific, Latin America and Caribbean, Northern Africa and Middle East, Southern Sub-Saharan Africa and Western Europe featured with 25–27 tests (out of 50). Most of these regions contain UMIC, with the exception of Western Europe. For Central Sub-Saharan Africa, Eastern Europe, Oceania and Western Africa all companies combined provided 21–24 tests. With the exception of Western Africa most of the countries in these regions are also UMIC. Eastern Sub-Saharan Africa, South Asia and Southeast Asia had the least number of total tests from all companies offered for prevalent diseases in the region with less than 20. South Asia had the lowest number with only 18. These regions have mostly LMIC and LIC.
Figure 2. (a+b): a shows the number of the most prevalent infectious diseases with PCR as diagnostics by DALYs from the GBD 2019 that the companies offer tests for. B is a world map showing the total number of tests from all companies offered for the 10 most frequent infectious diseases in the different regions of the world.
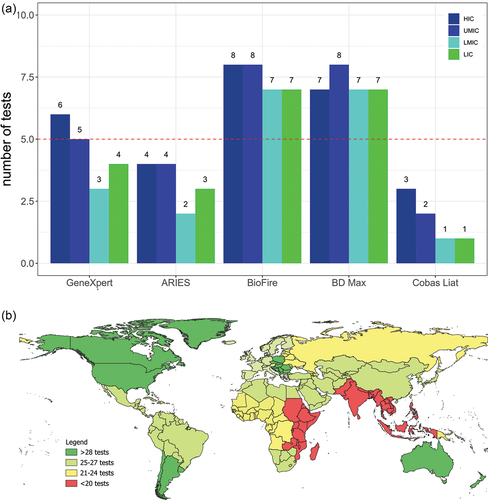
When adding up the total number of tests offered by all companies for the most frequent infectious diseases by region, it is striking to see that most tests are offered for the regions in the world where mostly HIC and UMIC are located. For South Asia, South East Asia and Eastern Sub-Saharan Africa there were almost 10 tests less offered than in HIC and UMIC, although LMIC and LIC are disproportionally affected by infectious diseases [Citation61] while being home to a big part of the world’s population. This is reflected in terms of total number of DALYs of the 10 most prevalent infectious diseases that can be diagnosed by PCR for the different world bank regions. For HIC, the total number is 6.05 million, for UMIC it is 31.3 million, for LMIC it is 132 million and for LIC it is 69 million, see supplementary table 3.
As seen in , of the 10 diseases with the highest burden, five of them were present in all of the regions, namely S. pneumococcus, HIV/AIDS, Tuberculosis, rotavirus, and RSV. Only one company, BioMerieux for S. pneumococcus and Cepheid for HIV/AIDS has a test on their menu. For Tuberculosis, Cepheid and BD offer a test in their portfolio and for rotavirus BioMerieux and BD. The only pathogen for which all companies offer a test is RSV. Varicella and herpes zoster virus (VZV) and C. difficile only were present in HIC list of the 10 most frequent diseases. Biomerieux offers a test for VZV, while all companies had a test for C. difficile. For the pathogens occurring in the 10 most prevalent infectious diseases in HIC and UMIC, all companies had a test for Influenza in their portfolio and BioMerieux and BD for Campylobacter. Norovirus was the only pathogen featured in the list of HIC, UMIC and LIC, and all companies except Roche Liat included it in one of their panels. Three companies (Luminex/Diasorin, BioMerieux, and BD) have a test for whooping cough, which was prevalent in UMIC and LMIC. For Shigella, important in UMIC, LMIC, and LIC, only two companies (BioMerieux and BD) offered a test in their portfolios. Three pathogens (adenovirus, S. typhi and cholera) are present only in LMIC. For adenovirus and cholera BioMerieux and BD offer tests, but no company offered a test for S. typhi (BD and BioMerieux include Salmonella spp. in their panel but it was only validated with S. typhimurium or S. enterica). Cryptosporidium and measles were only featured in the 10 most prevalent infectious diseases in LIC. BioMerieux and BD Max panels include a test for Cryptosporidium, but no company includes a test for measles.
Table 3. The most prevalent infectious diseases that can be diagnosed by PCR according to the regions where they occur.
Other than the vertically funded tuberculosis and HIV for LMIC, Cepheid only offers RSV and norovirus, two diseases that are very prevalent in HIC settings as well. Despite BD and Cepheid having a similar number of offered pathogens (35 and 24, respectively), their diagnostic reach in LMIC and LIC settings significantly varies. As seen in BD stands out by being able to offer 7 out of 10 of the tests required to diagnose the 10 most common diseases, while Cepheid lags behind with a coverage of only 2–3. Interestingly, BD Max manages to achieve a coverage of pathogens that closely rivals BioMerieux, despite having 100 fewer pathogens in their repertoire. Luminex/Diasorin and Cepheid consistently offer fewer pathogens across the board, particularly in the context of LMIC and LIC. This is a noteworthy contrast, particularly considering the widespread presence of Cepheid’s GeneXpert technology in LMIC and LIC settings. Probably due to GeneXpert’s market dominance, no other company offers tests for HIV and for TB only BD. Notably, only BD offers a test for S. pneumoniae, although it features highly in all regions.
The GBD 2019 does not contain SARS-Covid 2, for which all companies offer panels together with other respiratory infections such as RSV and Influenza A/B. Furthermore, it also has no data for AMR, for which all but Roche Liat offer panels.
Most companies focus on infectious diseases of primary importance to HIC, such as respiratory infections or hospital acquired infections. Although these diseases also occur in LMIC and are important to diagnose, it is clear that only few S2R platforms offer tests for diseases occurring mainly in LMIC, where there is a clear need for diagnostic capacity [Citation53]. Different mechanisms need to be applied to incentivize companies to develop tests for pathogens prevalent in LMIC. Cepheid’s success story with the GeneXpert for TB and HIV has shown that incentivizing companies with subsidies works [Citation62]. Another mechanism could be for funders, governments, or global health organizations (such as WHO, FIND, Gavi, and Cepi) to agree on purchasing a certain number of tests after development, similar to what has happened for COVID-19 vaccines during the pandemic, as an instrument to de-risk private investments. However, provisions for sustainability have to be made. The West Africa Ebola outbreak has spurred the development of diagnostic tests for Ebola Virus diagnostics, resulting in the FDA and/or WHO approval of 14 tests for emergency use, but a few years later only 5 were still available for an outbreak inthe Democratic Republic of Congo (DRC) in 2018 [Citation63]. During an outbreak or epidemic a lot of funding goes into the development of new therapeutics, vaccines and also diagnostics, but once funding is drying up with the end of the emergency, only few products can sustain themselves on a commercial market [Citation35]. Another caveat is that diseases such as TB or HIV have vertical programs with strong donor funding. A lot of diseases do not get this kind of attention and sustained donor funding, making it difficult for subsidy schemes. Even after excluding publications of TB and HIV to see what other tests without vertical programs were used from GeneXpert, more than half (35 out of 63) Cepheid studies were done with subsidized tests, i.e. for Ebola [Citation52,Citation64–66], SARS-CoV-2 [Citation67–71], CT/NG [Citation72–77], HPV [Citation78], HCV [Citation79–81] and HBC [Citation82]. Without subsidies test prices are too high for most low-resource settings and companies cannot build a sustainable revenue model by only selling to these. However, where substantial public investments in developing a health technology are made, it is important to examine whether the public sector has received adequate returns for its investments [Citation62]. A highly prominent case of this is the development of the GeneXpert, where one study estimates a public investment of over $252 million and reduced prices for some cartridges for LMIC have been negotiated, however for most LMIC these prices are still not affordable [Citation62]. As of September 2023 Cepheid announced a reduction in price for Xpert MTB/RIF test to 7.79US$ for high-burden countries [Citation83], but this falls short of the 5.0US$ price requested by MSF and only applies to this specific test [Citation53]. It is estimated that the production cost of a cartridge is only 2.95–4.64US$ per cartridge. Negotiated preferential prices also apply to only a few of the tests offered by Cepheid, whereas all Xpert cartridges use the same technology developed in part with public funding. Equitable access could be assured by giving more rights to the public sector in terms of intellectual property and pricing [Citation62].
2.3. Status on S2R usage in LMIC
A literature search for the implementation of S2R in LMIC was performed on PubMed with no time or language restrictions. To limit the search to LMIC settings, following search was used: ((((((((((((((LMIC) OR (LIC)) OR (Africa)) OR (Asia)) OR (‘latin america’)) OR (‘south america’)) OR (low income populations[MeSH Terms])) OR (africa[MeSH Terms])) OR (africa south of the sahara[MeSH Terms])) OR (‘south-east Asia’),‘LMIC’[All Fields] OR ‘LIC’[All Fields] OR (‘africa’[MeSH Terms] OR ‘africa’[All Fields] OR ‘africa s’[All Fields] OR ‘africas’[All Fields]) OR (‘asia’[MeSH Terms] OR ‘asia’[All Fields]) OR (‘latin america’[MeSH Terms] OR (‘latin’[All Fields] AND ‘america’[All Fields]) OR ‘latin america’[All Fields]) OR (‘south america’[MeSH Terms] OR (‘south’[All Fields] AND ‘america’[All Fields]) OR ‘south america’[All Fields]) OR ‘poverty’[MeSH Terms] OR ‘africa’[MeSH Terms] OR ‘africa south of the sahara’[MeSH Terms] OR (‘asia, southeastern’[MeSH Terms] OR (‘asia’[All Fields] AND ‘southeastern’[All Fields]) OR ‘southeastern asia’[All Fields] OR (‘south’[All Fields] AND ‘east’[All Fields] AND ‘asia’[All Fields]) OR ‘south east asia’[All Fields])))
Since one of the machines is called ARIES and a sheep breed called ‘Ovis Aries’ exists, anything related to ‘Ovis Aries’ or domestic sheep was excluded from the search with the following: (‘Ovis aries’) OR (‘O. aries’),‘sheep, domestic’[MeSH Terms] OR (‘sheep’[All Fields] AND ‘domestic’[All Fields]) OR ‘domestic sheep’[All Fields] OR (‘ovis’[All Fields] AND ‘aries’[All Fields]) OR ‘ovis aries’[All Fields] OR (‘o’[All Fields] AND ‘aries’[All Fields])”
‘BioFire,’ ‘ARIES,’ ‘BD Max’ and ‘Cobas Liat’ were searched in [All Fields] with the LMIC algorithm with the following:
For GeneXpert, the following search terms were used: (((‘Genexpert’) OR (‘Xpert’) OR (‘Cepheid’)))) NOT ((((tuberculosis) OR (‘M. tuberculosis’) OR (MDR) OR (‘multi-resistant tuberculosis’) OR (TB) OR (‘MTB RIF’))) OR ((((HIV) OR (AIDS) OR (‘HIV’ AND ‘early infant diagnosis’) OR (‘HIV’ AND ‘quantitative PCR’))))))) NOT (cancer)
Inclusion and Exclusion Criteria for literature search of S2R platform use in LMIC.
LMIC were defined as per the world bank definitions [October 2023] [Citation84] and studies performed in countries not in agreement with these definitions were excluded. Papers not related to the S2R platforms were also excluded.
When searching for the term ‘Cobas Liat’ and the LMIC algorithm on PubMed, only six articles were found. These were subsequently excluded from consideration due to the fact that the research conducted in these articles was carried out in countries classified as HIC (the UAE, Japan, South Korea, and Taiwan).
In the search for ‘ARIES’ along with the LMIC algorithm, a total of 56 publications were identified. All these publications had to be disregarded, as none of them were found to be associated with the ARIES S2R instrument.
The combined use of ‘BD Max’ along with the LMIC algorithm yielded a total of 14 research papers. Four of these papers were excluded from consideration due to the fact that the research had been conducted in countries that do not fall within the LMIC category, specifically France, Japan, Singapore, and Taiwan.
Apart from GeneXpert, the BioFire Assays by BioMérieux yielded the highest number of search results when used in conjunction with the LMIC algorithm. In total 66 papers were identified, out of which 39 were excluded. 13 were omitted because the BioFire analysis were not done in countries categorized as LMIC (Hong Kong, Japan, Taiwan, Israel, France, South Korea, Sweden) and 26 because the topic was not related to the BioFire assays.
When searching for GeneXpert in PubMed at the time of the search [17 October 2023] 5.913 publications are found, together with the LMIC algorithm this comes down to 1,788. Only publications related to infectious diseases were further considered. The validation and clinical utility of the use of GeneXpert with tuberculosis and HIV has been described extensively, so our research was focused on the other assays that GeneXpert offers and papers mentioning HIV or TB were excluded. In total 131 publications were retrieved, 68 were excluded for analysis, as their topic was not related to the use of GeneXpert, or the studies were not done in LMIC and 63 used for analysis.
Since our algorithm for the literature focused on Latin America, Sub-Saharan Africa, South Asia, and Southeast Asia, publications from UMIC in Europe might have been missed. Also, some publications from HIC but done in LRS (such as remote places in Canada or Australia) might not have been included, as this is out of the scope of this narrative review. Since cancer was excluded from the literature research, we might have excluded some publications from GeneXpert with HPV, a viral disease linked to cancer.
All data analysis and graphical work was done with Microsoft excel and R Studio. (a+b+c+d) A total of 103 publications were included in the analysis. For the different S2R platforms, this came to 10 publications with BD, 29 with BioFire and 60 with Cepheid, and 3 publications with analyzing Cepheid and BioFire (2.9%) and 1 GeneXpert and BD Max (1%). Even without including publications regarding TB, HIV, or cancer, over half of the total publications were using GeneXpert (58.3%). BioFire was used by 28.2% and BD Max by 9.7% (),
Figure 3. (a+b+c+d): a percentage of studies per company. b percentage of studies per income region. c percentage of studies per study type. d number of publications per country.
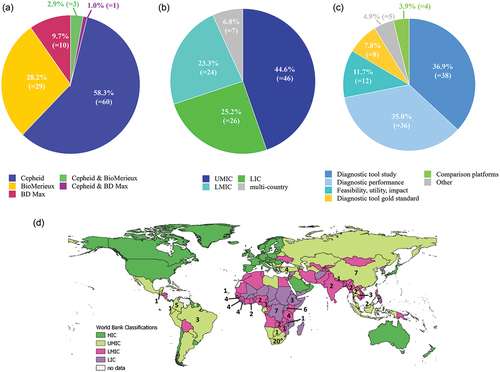
As seen in , almost half of the publications were done in UMIC (46 studies, 44.7%), with most studies coming from South Africa (20 studies). LMIC and LIC were almost equally represented with 24 studies (23.3%) and 26 studies (25.2%), respectively. Seven (6.8%) were multi-country studies. Almost half of the studies done in LIC (12 out of 26) and 5 out of 24 in LMIC, were Ebola studies. Most of them were published after the West-Africa Ebola outbreak in 2013–2016. Others are from ongoing smaller outbreaks in DRC, accounting for five out of the seven studies performed in DRC (other two were on mpox and SARS-CoV-2).
In almost half of the publications (44.7%), the S2R platforms were used as diagnostic tool in the studies, be it as purely diagnostic tool to investigate the epidemiology (36.9%), or as gold standard to evaluate the performance of other diagnostic tools or the adequacy of a new sample type (7.8%) (). Some of the papers used it as diagnostic tool in a study but at the same time assessed the diagnostic performance [Citation85], compared two different sample types for easier sample collection [Citation86,Citation87] or to another S2R platform [Citation88]. Validating different sample types that might be easier to obtain in LRS, such as capillary versus venous blood [Citation86] or rectal swabs instead of stool [Citation87] are crucial and more studies should be done on this. This is also shown with the C. trachomatis tests the Cepheid and BD offer as part of their STI panels. This test is only validated with samples for genital chlamydial infections, such as urine, vaginal, endocervical or rectal swabs [Citation89]. The GeneXpert is also validated on a non-genital related sample, a pharyngeal swab [Citation90]. However, in LMIC it is mostly used for the diagnosis of trachoma which requires ocular swabs [Citation72–77]. Even though these companies included the causing pathogen in their panels, they only validated it for the disease presentation in HIC as genital infection but not for eye infections, causing irreversible visual impairments or blindness in about 19 million people [Citation91].
Diagnostic performance studies constituted 35.0% of studies. 11 of the diagnostic performance studies also evaluated parameters beyond the accuracy of the S2R platform. The performance under field conditions was assessed by three studies [Citation79,Citation82,Citation92] while two studies [Citation93,Citation94] additionally looked at TAT and costs, whereas one only at TAT [Citation71]. Furthermore, one study [Citation95] compared two S2R platforms against each other and the reference standard. Only one study looked at the clinical utility [Citation77] and one additionally on the impact on antibiotic treatment prescription [Citation96]. In one paper, besides determining its diagnostic performance, the authors used it in a training [Citation97]. In one case, the S2R platform was compared to other diagnostics with the appearance of a new strain and potential mismatches [Citation98]. Diagnostic performance studies done in different regions in the world are crucial as most companies just validate the test in US-American or European setting [Citation12,Citation28], but different strains of pathogens exist and might affect the sensitivity and specificity of the S2R platform. Furthermore, diagnostic accuracy might be overstated by the manufacturer and should be tested independently, especially with strains more common in LMIC [Citation12]. This was investigated by a study about Ebola in DRC [Citation98] and one in Cambodia with HCV as the genotype 6 (GT) is dominant there, but most of the validations in HIC included GT1 and GT3 HCV [Citation79].
3.9% of the studies compared different S2Rs to each other [Citation95,Citation99–101]. 4.9% of the S2R were mentioned in other publications, such as reports, descriptive analysis or reviews [Citation102–106].
Only 11.7% (12 publications) were studies done beyond the diagnostic accuracy and focused on implementation criteria, such as feasibility, utility, and impact. The most frequently investigated parameters were TAT [Citation52,Citation66,Citation107,Citation108], impact on antimicrobial therapeutic decisions [Citation107–110], cost [Citation52,Citation80] and throughput [Citation52,Citation66]. Only one paper each was found to investigate provider satisfaction [Citation108], compare four different S2R platforms on their feasibility, ease-of-use and operational characteristics [Citation111] and feasibility, acceptability, effectiveness, and cost-effectiveness [Citation80]. Other research was done on impact of sample storage time and temperature on the performance [Citation112], feasibility in a simplified decentralized testing model [Citation81], implementation [Citation64] with the identification of logistic and safety issues [Citation66] and impact of the implementation on the case management, outbreak response and general strengthening of the laboratory system [Citation65]. More implementation research is needed to identify barriers to meaningful implementation of these machines and how to best remove them [Citation30,Citation32]. Implementation strategies cannot just be copied from HIC as the problems with lack of integration and service coordination with the hub-and-spoke model in LMIC is evident. There is a clear need for the use and reporting of implementation frameworks when evaluating programs for S2R platforms to expedite the conversion of research into policy and practice [Citation32].
2.4. Conclusion
S2R platforms are suitable for application in LMIC, however more studies that investigate parameters beyond diagnostic performance are needed, such as studies that look into cost-effectiveness, but also impact on providers, patients, and public health, over a longer period of time. Companies should have LMICs in mind when developing and validating their tests, to look into different sample types and into consumables stabilities with higher temperatures. They should also take more responsibility for waste disposal and invest in greener and less toxic technology to ease the waste problem in LMICs. Furthermore, it is essential to develop tests for pathogens that are occurring in LMIC. Alternative funding schemes are needed for diseases with no vertical program. The supply chain and service are major issues, which could be solved by identifying or creating production infrastructure in LMICs (similar to generic medicine market from Indian producers and recent investments in vaccine production capacity in Sub-Saharan Africa). Local companies should be encouraged and also benefit from funding schemes to bring products to market and publish validations. The production in LMIC might also help with supply chain issues as faced during the COVID-19 pandemic. S2R platforms are not meant to replace all conventional PCR diagnostic capacity, and options should always be explored if implementation of diagnostic capacity is not possible in the setting as this is still cheaper and more flexible.
3. Expert opinion
The COVID-19 pandemic accelerated the development and implementation of PCR, S2R platforms and even pathogen genomics analysis around the world, and this is likely to continue in the future. However, the implementation of S2R platforms in LMIC is limited due to high costs, limited access to service and the test menu being mainly directed for HIC. Manufacturers could implement smaller and easier changes, such as reagents with longer shelve life and validation of storage conditions beyond ‘room temperature.’ Different sample types could also be explored by manufacturers, either sample types important for disease presentation in LMIC or sample types that are easier to obtain, such as capillary blood. Bigger changes such as adapting machines to temperature, dust, and humidity or inclusion of priority pathogens in LMIC or lowering costs could be more complicated to achieve. Manufacturers should be held accountable for equitable access to their products, especially if they receive public funding for the development. Open systems where laboratories in LMIC could develop and validate tests meaningful to their settings could also be a way to increase the test portfolio. Funding or subsidizing companies for manufacturing in LMIC could be another way to achieve this. Technological developments, such as microfluidics could lower the costs of the cartridges. New and smaller devices with shorter running times and less infrastructure requirements, such as battery powered devices, could ease the implementation in LMIC.
More studies in LMIC on cost-effectiveness and parameters beyond the diagnostic performance are urgently needed to assess the clinical utility and public health impact, and to identify bottlenecks during the implementation. There should be ‘structured implementation’ studies so that comparison between settings in different countries is easier. Strengthening of health systems is vital as the best diagnostic test is useless if there is no linkage to care and appropriate treatments for diagnosed diseases. Digitalization is also essential for better communication of results to patients and public health authorities so that patients can get appropriate care and treatment, and public health authorities can act quickly in events such as epidemics.
New technologies beyond the melting curve or probe hydrolysis assay are promising to extend the possibilities of molecular diagnostics. These include LAMP, CRISPR-Cas and Ion Sensitive Field Effect Transistors (ISFET), amongst many others. They can help to achieve shorter running times, lower costs, and smaller machines requiring less infrastructure.
More products should come from emerging countries such as India, China, Brazil, and South Africa, as they can start to cater to their own needs, which has already been shown with drug production in India.
Furthermore, technologies beyond PCR will gain importance such as pathogen genome sequencing. Depending on the strategy, it allows unbiased diagnostics, enabling the detection of rare or new pathogens and unknown mutations, facilitating determination of new resistances and strains. In the future, it might be incorporated in clinical algorithms for samples negative to common pathogens, but if the price decreases enough, it might become the diagnostic of choice. Simplification and automatization will allow its application also in LMIC, similar to PCR and S2R platforms.
Article highlights
The complexity that comes with molecular diagnostics can be a hurdle for implementation, especially in Low- and Middle-Income Countries (LMIC).
Sample-to-Result (S2R) platforms have several advantages, such as ease-of-use, fast turnaround time (TAT), limited requirements on infrastructure and technician training.
Several shortcomings limit their implementation in LMIC, such as costs, validation of equipment and reagents in environments common in LMIC, waste disposal, distribution, customer service, and platform maintenance.
Test menus of S2R platforms are tailored to meet the need of infectious diseases prevalent in HIC, although the number of Disability-Adjusted Life Years (DALYs) of the 10 most frequent infectious diseases are a lot higher in LMIC or Low-Income Countries (LIC) than in Upper Middle-Income Countries (UMIC) and High-Income Countries (HIC)
With the exception of GeneXpert, there are not a lot of publications of S2R applications in LMIC. Literature reports mostly on their use as diagnostic tool in studies or on the technical evaluation of the platform, but very few studies report about implementation or operational characteristics and not in a unified way.
Declaration of interest
C Onwuchekwa is an employee of p-95. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
Supplemental Material
Download MS Word (272.3 KB)Acknowledgments
The authors acknowledge the use of ChatGPT (Version 3.5), an OpenAI software to rewrite some of the author’s sentences for better clarity and grammar. The authors are grateful to Stijn Roge and Samant Nagraj for critically reading this manuscript.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/14737159.2024.2353690
Additional information
Funding
References
- Hay Burgess DC, Wasserman J, Dahl CA. Global health diagnostics: foreword. Nature. 2006;444(S1):1–2. doi: 10.1038/nature05440
- Fleming KA, Horton S, Wilson ML, et al. The Lancet commission on diagnostics: transforming access to diagnostics. Lancet (London, England). 2021;398:1997–2050. doi: 10.1016/S0140-6736(21)00673-5.
- Daar AS, Thorsteinsdóttir H, Martin DK, et al. Top Ten biotechnologies for improving health in developing countries. Nature Genet. 2002;32(2):229–232. doi: 10.1038/ng1002-229
- Jani IV, Peter TF. How point-of-care testing could drive innovation in global health. N Engl J Med. 2013;368(24):2319–2324. doi: 10.1056/NEJMsb1214197
- Drain PK, Hyle EP, Noubary F, et al. Diagnostic point-of-care tests in resource-limited settings. Lancet Infect Dis. 2014;14(3):239–249. doi: 10.1016/S1473-3099(13)70250-0
- Mabey D, Peeling RW, Ustianowski A, et al. Diagnostics for the developing world. Nature Rev Microbiol. 2004;2(3):231–240. doi: 10.1038/nrmicro841
- Schroeder LF, Pai M. A list to cement the rightful place of diagnostics in health care. J Clin Microbiol. 2018;56(10). doi: 10.1128/JCM.01137-18
- Proesmans S, Katshongo F, Milambu J, et al. Dengue and chikungunya among outpatients with acute undifferentiated fever in Kinshasa, democratic Republic of Congo: a cross-sectional study. PLOS Negl Trop Dis. 2019;13(9):1–16. doi: 10.1371/journal.pntd.0007047
- Carrillo-Hernández MY, Ruiz-Saenz J, Villamizar LJ, et al. Co-circulation and simultaneous co-infection of dengue, chikungunya, and zika viruses in patients with febrile syndrome at the Colombian-Venezuelan border. BMC Infect Dis. 2018;18(1):1–12. doi: 10.1186/s12879-018-2976-1
- Nedjadi T, El-Kafrawy S, Sohrab SS, et al. Tackling dengue fever: Current status and challenges. Virol J. 2015;12(1):1–11. doi: 10.1186/s12985-015-0444-8
- Pai M, Ghiasi M, Pant Pai N. Point-of-care diagnostic testing in global health: what is the point? Microbe. 2015;10(3):103–107. doi: 10.1128/microbe.10.103.1
- Kosack CS, Page A-L, Klatser PR. A Guide to aid the selection of diagnostic tests. Bullet World Health Organ. 2017;95(9):639–645. doi: 10.2471/BLT.16.187468
- World Health Organization. A global strategy to eliminate yellow fever epidemics (EYE) 2017-2026. 2018 Jan 21. Licence: CC BY-NC-SA 3.0 IGO. https://www.who.int/publications/i/item/9789241513661
- Mansuy J-M, Lhomme S, Cazabat M, et al. Detection of Zika, dengue and chikungunya viruses using single-reaction multiplex real-time RT-PCR. Diagn Microbiol Infect Dis. 2018;92(4):284–287. doi: 10.1016/j.diagmicrobio.2018.06.019
- Semret M, Ndao M, Jacobs J, et al. Point-of-care and point-of-‘can’: leveraging reference-laboratory capacity for integrated diagnosis of fever syndromes in the tropics. Clin Microbiol Infect. 2018;24:836–844. doi: 10.1016/j.cmi.2018.03.044
- Kost GJ. Molecular and point-of-care diagnostics for Ebola and new threats: national POCT policy and guidelines will stop epidemics. Expert Rev Mol Diagn. 2018;18(7):657–673. doi: 10.1080/14737159.2018.1491793
- Altindiş M, Kahraman Kilbaş EP. Managing viral emerging infectious diseases via Current and future molecular diagnostics. Diagnostics (Basel). 2023;13(8):1421. doi: 10.3390/diagnostics13081421
- Dwivedi S, Purohit P, Misra R, et al. Diseases and molecular diagnostics: a step closer to precision medicine. Ind J Clin Biochem. 2017;32(4):374–398. doi: 10.1007/s12291-017-0688-8
- Schmitz JE, Stratton CW, Persing DH, et al. Forty years of molecular diagnostics for infectious diseases. J Clin Microbiol. 2022;60(10):e0244621. doi: 10.1128/jcm.02446-21
- Guzmán MG, Kourı́ G. Dengue diagnosis, advances and challenges. Inter J Infect Dis. 2004;8(2):69–80. doi: 10.1016/j.ijid.2003.03.003
- Beal SG, Assarzadegan N, Rand KH. Sample-to-result molecular infectious disease assays: clinical implications, limitations and potential, expert review of molecular diagnostics 16. Expert Rev Mol Diagn. 2016;16(3):323–341. doi: 10.1586/14737159.2016.1134325
- Perkins MD, Kessel M. What Ebola tells us about outbreak diagnostic readiness. Nature Biotechnol. 2015;33(5):464–469. doi: 10.1038/nbt.3215
- WHO. A look inside laboratories responding to COVID-19 [cited 2023 Nov 26] Available from: https://www.who.int/news-room/feature-stories/detail/a-look-inside-laboratories-responding-to-covid-19
- Okeke IN, Ihekweazu C. The importance of molecular diagnostics for infectious diseases in low-resource settings. Nature Rev Microbiol. 2021;19(9):547–548. doi: 10.1038/s41579-021-00598-5
- WHO. Benin boosts COVID-19 response with increased testing [cited 2023 Nov 26]. Available from: https://www.afro.who.int/news/benin-boosts-covid-19-response-increased-testing
- McKinsey & Company. COVID-19 and in vitro diagnostics: new market forces at play [cited 2023 Nov 26]. Available from: https://www.mckinsey.com/industries/life-sciences/our-insights/covid-19-and-in-vitro-diagnostics-new-market-forces-at-play
- WHO. Increased testing capacity, essential step in fighting COVID-19 [cited 2023 Nov 26]. Available from: https://www.who.int/bangladesh/news/detail/08-10-2020-increased-testing-capacity-essential-step-in-fighting-covid-19
- Sharma S, Crawley A, O’Kennedy R. Strategies for overcoming challenges for decentralised diagnostics in resource-limited and catastrophe settings. Expert Rev Mol Diagn. 2017;17(2):109–118. doi: 10.1080/14737159.2017.1273773
- WHO Global TB Programme. Xpert MTB/RIF implementation manual: technical and operational ‘how-to’: practical considerations. preprint.2014 [cited 2023 Jun 13]. Available from: https://www.ncbi.nlm.nih.gov/books/NBK254323/pdf/Bookshelf_NBK254323.pdf
- Albert H, Nathavitharana RR, Isaacs C, et al. Development, roll-out and impact of xpert MTB/RIF for tuberculosis: what lessons have we learnt and how can we do better? Eur Respir J. 2016;48(2):516–525. doi: 10.1183/13993003.00543-2016
- Beall SG, Cantera J, Diaz MH, et al. Performance and workflow assessment of six nucleic acid extraction technologies for use in resource limited settings. PLOS ONE. 2019;14(4):e0215753. doi: 10.1371/journal.pone.0215753
- Brown S, Leavy JE, Jancey J. Implementation of GeneXpert for TB testing in low-and middle-income countries: a systematic review. Global Health. 2021;9(3):698–710. doi: 10.9745/GHSP-D-21-00121
- Roche Molecular Diagnostics, Cobas Liat System: lab in a tube. Available from: http://sub.chimei.org.tw/55399/images/PDF/06_education/01_class/20170926_cobasz.pdf
- Lehe JD, Sitoe NE, Tobaiwa O, et al. Evaluating operational specifications of point-of-care diagnostic tests: a standardized scorecard. PLOS ONE. 2012;7(10):e47459. doi: 10.1371/journal.pone.0047459
- Emperador DM, Mazzola LT, Kelly-Cirino C. An open-source molecular diagnostic platform approach for outbreak and epidemic preparedness. Afr J Lab Med. 2020;9(2):1017. doi: 10.4102/ajlm.v9i2.1017
- Her T, Schutzbank TE. Evaluation of the luminex ARIES® system for the detection and quantification of BK virus (BKV) DNA in plasma samples from kidney transplant recipients. Diagn Microbiol Infect Dis. 2019;94(2):129–134. doi: 10.1016/j.diagmicrobio.2018.12.004
- Liu B, Totten M, Nematollahi S, et al. Development and evaluation of a fully automated molecular assay targeting the mitochondrial small subunit RRNA gene for the detection of pneumocystis jirovecii in bronchoalveolar lavage fluid specimens. J Mol Diagn. 2020;22(12):1482–1493. doi: 10.1016/j.jmoldx.2020.10.003
- Frans G, Beuselinck K, Peeters B, et al. Migrating a lab-developed MERS-CoV real-time PCR to 3 “sample to result” systems: experiences on optimization and validation. Diagn Microbiol Infect Dis. 2019;94(4):349–354. doi: 10.1016/j.diagmicrobio.2019.02.006
- Wolf LA, Marimuthu S, Summersgill JT. Detection of Ehrlichia Spp. And Anaplasma Phagocytophilum in whole blood specimens using a duplex real-time PCR assay on the ARIES Instrument. Ticks Tick-Borne Dis. 2020;11(3):101387. doi: 10.1016/j.ttbdis.2020.101387
- Certest. Viasure: complete solution [cited 2023 Oct 20]. Available from: https://www.certest.es/viasure/
- WHO. Safe management of wastes from health-care activities [cited 2023 Oct 24]. Available from: https://iris.who.int/bitstream/handle/10665/85349/9789241548564_eng.pdf;jsessionid=82A5E5C690C67FC417FC5568300525EB?sequence=1
- Street A, Vernooij E, Rogers MH. Diagnostic waste: whose responsibility? Globalization Health. 2022;18(1):30. doi: 10.1186/s12992-022-00823-7
- Odhiambo CO, Mataka A, and Kassa G, et al. Managing laboratory waste from HIV-Related molecular testing: lessons learned from African Countries. J Hazard Mater Lett. 2021, None. 2:100030. doi: 10.1016/j.hazl.2021.100030
- BD. REACH - AFA021-01 OPnEO chemical safety report public: chemical safety report [cited 2023 Oct 18]. Available from: https://consultations.hse.gov.uk/crd-reach/afa021-01-opeo/supporting_documents/REACH%20%20AFA02101%20OPnEO%20Chemical%20Safety%20Report%20Public.pdf
- The Global Fund. Technical brief: avoidance, reduction and sage management of health care waste: allocation period 2023-2025 [cited 2023 Oct 23]. Available from: https://www.theglobalfund.org/media/9356/core_healthcarewastemanagement_technicalbrief_en.pdf
- Unitaid. Guidance note on product selection-facility upgrades and sample transportation [cited 2023 Oct 23]. Available from: https://www.childrenandaids.org/sites/default/files/2019-08/Guidance%20Note%20on%20Product%20Selection-Facility%20Upgrades%20and%20Sample%20Transportation.pdf
- Celis JE, Espejo W, Paredes-Osses E, et al. Plastic residues produced with confirmatory testing for COVID-19: classification, quantification, fate, and impacts on human health. Sci Total Environ. 2021;760:144167. doi: 10.1016/j.scitotenv.2020.144167
- WHO. Progress on WASH in health care facilities 2000-2021: special focus on WASH and infection prevention and control (IPC) [cited 2023 Oct 24]. Available from: https://data.unicef.org/resources/jmp-wash-in-health-care-facilities-2022/
- Schneidman M, Dacombe RJ, Carter J. Laboratory professionals in Africa: the backbone of quality diagnostics.
- Africa Medical Supplies Platform [cited 2023 Oct 18]. Available from: https://amsp.africa/
- Bårnes GK, Gudina EK, Berhane M, et al. New molecular tools for meningitis diagnostics in Ethiopia – a necessary step towards improving antimicrobial prescription. BMC Infect Dis. 2018;18(1):684. doi: 10.1186/s12879-018-3589-4
- Schuh AJ, Kyondo J, Graziano J, et al. Rapid establishment of a Frontline Field Laboratory in response to an imported outbreak of Ebola virus disease in Western Uganda, June 2019. PLOS negl trop dis. 2021;15(12):e0009967. doi: 10.1371/journal.pntd.0009967
- Cantera JL, White H, Diaz MH, et al. Assessment of eight nucleic acid amplification technologies for potential use to detect infectious agents in low-resource settings. PLOS ONE. 2019;14(4):e0215756. doi: 10.1371/journal.pone.0215756
- Abdurrahman ST, Emenyonu N, Obasanya OJ, et al. The hidden costs of installing xpert machines in a tuberculosis High-Burden Country: experiences from Nigeria. Pan Afr Med J. 2014;18:277. doi: 10.11604/pamj.2014.18.277.3906
- Nkengasong J. Let Africa into the market for COVID-19 diagnostics. Nature. 2020;580(7805):565–565. doi: 10.1038/d41586-020-01265-0
- National Health Laboratory Service. Annual performance plan: fiscal year: 2019/2020 [cited 2023 Oct 23]. Available from: https://www.nhls.ac.za/wp-content/uploads/2019/11/NHLS_APP_2019.pdf
- WHO. Health systems financing: the path to universal coverage [cited 2023 Oct 23]. Available from: https://iris.who.int/bitstream/handle/10665/44371/9789241564021_eng.pdf?sequence=1
- Barroy H, Sparkes S, Dale E, et al. Can low- and middle-income countries increase domestic fiscal space for health: a mixed-methods approach to assess possible sources of expansion. Health Syst Reform. 2018;4(3):214–226. doi: 10.1080/23288604.2018.1441620
- Perry L, Malkin R. Effectiveness of medical equipment donations to improve health systems: how much medical equipment is broken in the developing world? Med Biol Eng Comput. 2011;49(7):719–722. doi: 10.1007/s11517-011-0786-3
- IHME. GBD Results: Explore Results From The 2019 Global Burden Of Disease (GBD) Study [cited 2023 Oct 4]. Available from: https://vizhub.healthdata.org/gbd-results/
- WHO. World health statistics 2022: monitoring health for the SDGs [cited 2023 Oct 20]. Available from: https://cdn.who.int/media/docs/default-source/gho-documents/world-health-statistic-reports/worldhealthstatistics_2022.pdf
- Gotham D, McKenna L, Deborggraeve S, et al. Public Investments in the development of GeneXpert molecular diagnostic technology. PLOS ONE. 2021;16(8):e0256883. doi: 10.1371/journal.pone.0256883
- Cnops L, de Smet B, Mbala-Kingebeni P, et al. Where are the Ebola diagnostics from last time? Nature. 2019;565(7740):419–421. doi: 10.1038/d41586-019-00212-y
- Katawera V, Kohar H, Mahmoud N, et al. Enhancing laboratory capacity during Ebola virus disease (EVD) heightened surveillance in Liberia: lessons learned and recommendations. Pan Afr Med J. 2019;33:8. doi: 10.11604/pamj.supp.2019.33.2.17366
- Raftery P, Condell O, Wasunna C, et al. Establishing Ebola virus disease (EVD) diagnostics using GeneXpert technology at a Mobile laboratory in Liberia: impact on outbreak response, case management and laboratory systems Strengthening, PLOS neglected tropical diseases 12. PLOS negl trop dis. 2018;12(1):e0006135. doi: 10.1371/journal.pntd.0006135
- van den Bergh R, Chaillet P, Sow MS, et al. Feasibility of Xpert Ebola assay in médecins sans frontières Ebola program, Guinea. Emerg Infect Dis. 2016;22(2):210–2106. doi: 10.3201/eid2202.151238
- Noble LD, Scott LE, Munir R, et al. Rapid evaluation of the Xpert® Xpress CoV-2 plus and Xpert® xpress CoV-2/Flu/RSV plus tests. Diagnostics (Basel). 2022;13:1–16.
- Makulo J-R, Mandina MN, Mbala PK, et al. SARS-CoV2 infection in symptomatic patients: interest of serological tests and predictors of mortality: experience of DR Congo. BMC Infect Dis. 2022;22(1):21. doi: 10.1186/s12879-021-07003-9
- Dorkenoo AM, Gbeasor-Komlanvi FA, Gbada K, et al. Prevalence of malaria and covid-19 in febrile patients in Lomé, Togo in 2020. Acta Parasit. 2022;67(3):1335–1342. doi: 10.1007/s11686-022-00586-6
- Chen JH-K, Yip CC-Y, Poon RW-S, et al. Evaluating the use of posterior oropharyngeal saliva in a point-of-care assay for the detection of SARS-CoV-2. Emerg Microbes Infect. 2020;9:1356–1359. doi: 10.1080/22221751.2020.1775133
- Wen D, Yang S, Li G, et al. Sample-to-answer and routine real-time RT-PCR: a comparison of different platforms for SARS-CoV-2 detection. J Mol Diagn. 2021;23(6):665–670. doi: 10.1016/j.jmoldx.2021.02.010
- Odonkor M, Naufal F, Munoz B, et al. Serology, infection, and clinical trachoma as tools in prevalence surveys for Re-emergence of trachoma in a formerly hyperendemic District, PLOS neglected tropical diseases 15. PLOS negl trop dis. 2021;15(4):e0009343. doi: 10.1371/journal.pntd.0009343
- West SK, Munoz B, Mkocha H, et al. The effect of Mass Drug Administration for Trachoma on antibodies to chlamydia trachomatis Pgp3 in children. Sci Rep. 2020;10(1):15225. doi: 10.1038/s41598-020-71833-x
- Burr SE, Hart J, Samikwa L, et al. Pgp3 seroprevalence and associations with active trachoma and Ocular Chlamydia Trachomatis Infection in Malawi: cross-sectional surveys in six evaluation units. PLOS Negl Trop Dis. 2019;13(10):e0007749. doi: 10.1371/journal.pntd.0007749
- Dize L, West S, Williams JA, et al. Comparison of the Abbott M2000 RealTime CT Assay and the Cepheid GeneXpert CT/NG Assay to the Roche Amplicor CT Assay for Detection of Chlamydia Trachomatis in ocular samples from Tanzania. J Clin Microbiol. 2013;51(5):1611–1613. doi: 10.1128/JCM.00519-13
- Senyonjo LG, Debrah O, Martin DL, et al. Serological and PCR-Based Markers of Ocular Chlamydia Trachomatis Transmission in northern Ghana after elimination of trachoma as a public health Problem, PLOS neglected tropical diseases 12. PLOS negl trop dis. 2018;12(12):e0007027. doi: 10.1371/journal.pntd.0007027
- Jenson A, Dize L, Mkocha H, et al. Field evaluation of the Cepheid GeneXpert Chlamydia Trachomatis assay for detection of infection in a Trachoma Endemic Community in Tanzania, PLOS neglected tropical diseases. PLOS negl trop dis. 2013;7(7):e2265. doi: 10.1371/journal.pntd.0002265
- Khoo SP, Shafii MKA, Bhoo-Pathy N, et al. Prevalence and sociodemographic correlates of Anogenital human papillomavirus (HPV) carriage in a cross-sectional, multi-ethnic, community-based Asian male population. PLOS ONE. 2021;16(1):e0245731. doi: 10.1371/journal.pone.0245731
- Iwamoto M, Calzia A, Dublineau A, et al. Field evaluation of GeneXpert® (cepheid) HCV performance for RNA quantification in a genotype 1 and 6 predominant patient population in Cambodia. J Viral Hepat. 2019;26(1):38–47. doi: 10.1111/jvh.13002
- Draper BL, Pedrana A, Howell J, et al. Decentralized, community-based hepatitis C point-of-care testing and direct-acting antiviral treatment for people who inject drugs and the General population in Myanmar: protocol for a feasibility study. JMIR Res Protoc. 2020;9(7):e16863. doi: 10.2196/16863
- Zhang M, O’Keefe D, Craig J, et al. Decentralised hepatitis C testing and treatment in rural Cambodia: evaluation of a simplified service Model integrated in an Existing Public Health System. Lancet Gastroenterol Hepatol. 2021;6(5):371–380. doi: 10.1016/S2468-1253(21)00012-1
- Woldemedihn GM, Rueegg CS, Desalegn H, et al. Validity of a point-of-care viral load test for hepatitis B in a low-income setting. J Virol Methods. 2021;289:114057. doi: 10.1016/j.jviromet.2020.114057
- MSF. MSF: tB Test Price Reduction by Cepheid and Danaher is an important step in the right direction [cited 2023 Oct 25]. Available from: https://msfsouthasia.org/msf-tb-test-price-reduction-by-cepheid-and-danaher-is-an-important-step-in-the-right-direction/
- World Bank. World bank country and lending groups. Available from: https://datahelpdesk.worldbank.org/knowledgebase/articles/906519-world-bank-country-and-lending-groups
- Putsathit P, Maneerattanaporn M, Piewngam P, et al. Prevalence and molecular epidemiology of Clostridium Difficile Infection in Thailand. New Microbes New Infect. 2017;15:27–32. doi: 10.1016/j.nmni.2016.10.004
- Leski TA, Taitt CR, Bangura U, et al. Comparison of capillary and venous blood for malaria detection using two PCR-Based assays in febrile patients in Sierra Leone. Malaria j. 2021;20(1):133. doi: 10.1186/s12936-021-03644-y
- Walker CR, Lechiile K, Mokomane M, et al. Evaluation of anatomically designed flocked rectal swabs for use with the BioFire FilmArray gastrointestinal panel for detection of enteric pathogens in children admitted to hospital with severe gastroenteritis. J Clin Microbiol. 2019;57(12). doi: 10.1128/JCM.00962-19
- Loevinsohn G, Hardick J, Sinywimaanzi P, et al. Respiratory pathogen diversity and Co-infections in Rural Zambia. Inter J Infect Dis. 2021;102:291–298. doi: 10.1016/j.ijid.2020.10.054
- CDRH. CT/GC/TV2 panel: 510(k) substantial equivalence determination decision memorandum assay and instrument combination template [cited 2023 Jul 31]. Available from: https://www.accessdata.fda.gov/cdrh_docs/reviews/K182692.pdf
- Cepheid. Xpert CTNG: uS ENGLISH package insert 301-0234, Rev K.Fm [cited 2023 Jun 26]. Available from: https://www.cepheid.com/content/dam/www-cepheid-com/documents/package-insert-files/Xpert-CTNG-US-ENGLISH-Package-Insert-301-0234–Rev-K.pdf
- WHO. Neglected tropical diseases [cited 2023 Oct 2] Available from: https://www.who.int/health-topics/neglected-tropical-diseases#tab=tab_1
- Semper AE, Broadhurst MJ, Richards J, et al. Performance of the GeneXpert Ebola assay for diagnosis of Ebola virus disease in Sierra Leone: a field evaluation study. PLOS Med. 2016;13(3):e1001980. doi: 10.1371/journal.pmed.1001980
- Ayebare A, Bebell LM, Bazira J, et al. Comparative Assessment of Methicillin resistant staphylococcus aureus diagnostic assays for use in resource-limited settings. BMC Microbiol. 2019;19(1):194. doi: 10.1186/s12866-019-1566-8
- Cury AP, Almeida Junior JN, Costa SF, et al. Diagnostic performance of the xpert carba-R™ assay directly from rectal swabs for active surveillance of carbapenemase-producing organisms in the largest Brazilian University hospital. J Microbiol Methods. 2020;171:105884. doi: 10.1016/j.mimet.2020.105884
- Singh S, Newton-Foot M, Nel P, et al. Comparison of commercial assays and two-step approach to detect clostridioides difficile in South Africa. Clinical Microbiology Reviews. 2022;11(1):1–6. doi: 10.4102/ajlm.v11i1.1809
- van der Westhuyzen M, Samodien N, Brink AJ, et al. Utility of the BioFire® FilmArray® Pneumonia panel plus assay for syndromic testing of lower respiratory tract infections in a Low/middle-income setting. JAC Antimicrob Resist. 2023;5(1):dlac139. doi: 10.1093/jacamr/dlac139
- Bentahir M, Barry MD, Koulemou K, et al. Providing on-site laboratory and biosafety just-in-time training inside a box-based laboratory during the west africa Ebola outbreak: Supporting better preparedness for future health emergencies. Int J Environ Res Public Health. 2022;19(18):11566. doi: 10.3390/ijerph191811566
- Mukadi-Bamuleka D, Bulabula-Penge J, Jacobs BKM, et al. Head-to-head comparison of diagnostic accuracy of four Ebola virus disease rapid diagnostic tests versus GeneXpert® in Eastern Democratic Republic of the Congo outbreaks: a prospective observational study. EBioMedicine. 2023;91:104568. doi: 10.1016/j.ebiom.2023.104568
- Zhan Z, Guo J, Xiao Y, et al. Comparison of BioFire FilmArray gastrointestinal panel versus Luminex XTAG gastrointestinal Pathogen panel (XTAG GPP) for Diarrheal Pathogen Detection in China. Inter J Infect Dis. 2020;99:414–420. doi: 10.1016/j.ijid.2020.08.020
- de Vos M, Scott L, David A, et al. Comparative analytical evaluation of four centralized platforms for the detection of mycobacterium tuberculosis complex and resistance to rifampicin and isoniazid. J Clin Microbiol. 2021;59(3). doi: 10.1128/JCM.02168-20
- Rajabally N, Kullin B, Ebrahim K, et al. A comparison of Clostridium Difficile diagnostic methods for identification of local strains in a South African centre. J Med Microbiol. 2016;65(4):320–327. doi: 10.1099/jmm.0.000231
- Inungu J, Iheduru-Anderson K, Odio OJ. Recurrent Ebola virus disease in the democratic Republic of Congo: update and challenges. 2019;6(4):502–513. doi: 10.3934/publichealth.2019.4.502
- Pan Z-Y, Wu Y-J, Zeng Y-X, et al. Pooled analysis of the accuracy of xpert Ebola assay for diagnosing Ebola virus infection. Bio Med Res Int. 2021;2021:5527505. doi: 10.1155/2021/5527505
- Kamulegeya R, Kateete DP, Bagaya BS, et al. Biobanking: strengthening Uganda’s Rapid Response to COVID-19 and other epidemics. Biopreserv Biobank. 2022;20:238–243. doi: 10.1089/bio.2021.0022
- Gudza-Mugabe M, Sithole K, Sisya L, et al. Zimbabwe’s emergency response to COVID-19: enhancing access and accelerating COVID-19 testing as the first line of defense against the COVID-19 pandemic. Front Public Health. 2022;10:871567. doi: 10.3389/fpubh.2022.871567
- Scott LE, Hsiao N-Y, Dor G, et al. How South Africa used national cycle threshold (ct) values to continuously monitor SARS-CoV-2 laboratory test quality. Diagnostics (Basel). 2023;13:1–13.
- Reddy K, Whitelaw A. Can the Xpert MRSA/SA BC assay be used as an antimicrobial stewardship tool? a prospective assay validation and descriptive impact assessment study in a South African setting. BMC Infect Dis. 2021;21(1):177. doi: 10.1186/s12879-021-05857-7
- Subramaniam A, Blanchard CT, Ngek ESN, et al. Prevalence of group B streptococcus anogenital colonization and feasibility of an intrapartum screening and antibiotic prophylaxis protocol in Cameroon, africa. Int J Gynaecol Obstet Off Organ Int Fed Gynaecol Obstet. 2019;146(2):238–243. doi: 10.1002/ijgo.12870
- Molina FJ, Botero LE, Isaza JP, et al. Diagnostic concordance between BioFire® FilmArray® Pneumonia Panel and culture in patients with COVID-19 Pneumonia admitted to intensive care units: the experience of the third wave in eight hospitals in Colombia. Crit Care. 2022;26(1):130. doi: 10.1186/s13054-022-04006-z
- Rule R, Paruk F, Becker P, et al. Clinical utility of the BioFire FilmArray blood culture identification panel in the adjustment of empiric antimicrobial therapy in the Critically Ill Septic Patient. PLOS ONE. 2021;16(7):e0254389. doi: 10.1371/journal.pone.0254389
- David A, de Vos M, Scott L, et al. Feasibility, ease-of-use, and Operational Characteristics of World Health Organization–recommended moderate-complexity automated nucleic acid amplification tests for the detection of tuberculosis and resistance to rifampicin and isoniazid. J Mol Diagn. 2023;25(1):46–56. doi: 10.1016/j.jmoldx.2022.10.001
- Maduna LD, Kock MM, Medina-Marino A, et al. Impact of specimen storage temperature and Time on the implementation of GeneXpert® testing for sexually transmitted infections in resource-constraint settings. J Microbiol Methods. 2019;165:105719. doi: 10.1016/j.mimet.2019.105719