Abstract
Retinoic acid is considered to be the active metabolite of retinol, able to control differentiation and proliferation of epithelia. Retinoic acid biosynthesis has been widely described with the implication of multiple enzymatic activities. However, our understanding of the cell biological function and regulation of this process is limited. In a recent study we evidenced that milk xanthine oxidase (E.C. 1.17.3.2.) is capable to oxidize all-trans-retinol bound to CRBP (holo-CRBP) to all-trans-retinaldehyde and then to all-trans-retinoic acid. To get further knowledge regarding this process we have evaluated the biosynthetic pathway of retinoic acid in a human mammary epithelial cell line (HMEC) in which xanthine dehydrogenase (E.C. 1.17.1.4.), the native form of xanthine oxidase, is expressed. Here we report the demonstration of a novel retinol oxidation pathway that in the HMEC cytoplasm directly conduces to retinoic acid. After isolation and immunoassay of the cytosolic protein showing retinol oxidizing activity we identified it with the well-known enzyme xanthine dehydrogenase. The NAD+ dependent retinol oxidation catalyzed by xanthine dehydrogenase is strictly dependent on cellular retinol binding proteins and is inhibited by oxypurinol. In this work, a new insight into the biological role of xanthine dehydrogenase is given.
Introduction
Vitamin A derivatives are vital for various physiological functions including embryonic development, reproduction, vision, and for the maintenance of various epithelia and the immune system. In particular, retinoic acid is considered to be the active metabolite of retinol, able to explain a variety of biological functions [Citation1] and preventive, curative effects in some pathological conditions Citation2-5. Retinoic acid biosynthesis has been widely described and many possible enzymatic activities for catalyzing the two-step oxidation of retinol to retinoic acid, as microsomal short-chain alcohol dehydrogenase/reductase (SRD) and retinol dehydrogenase cis-retinol/androgen dehydrogenase (CRAD), cytosolic alcohol (ADH) and aldehyde dehydrogenases (ALDH), xanthine oxidase (XO), and cytochrome P-450 isozymes have been identified Citation6-10. The prevailing description of this biosynthetic process [Citation11] is that retinol is first oxidized to retinaldehyde, which is then oxidized to retinoic acid [Citation6,Citation7,Citation10,Citation12], and the possible participation of protein binding these substrates has been considered [Citation6,Citation8,Citation13,Citation14]. The evidence that retinoids bounded to specific cellular retinoid binding proteins are a better substrate for some retinoid catalysis with respect to free retinoids has been well established [Citation14]. In particular, the study of Ottonello et al. [Citation14] carefully demonstrated the presence in calf liver of a cytosolic enzyme system utilizing CRBP-bound retinol, able to catalyze the direct oxidation of retinol to retinoic acid.
The interest in the way retinoic acid is produced and in the cooperation carried out by cellular retinoid binding proteins(CRBP(s)) is magnified in the case of tissues characterized by a functional dynamism as the breast tissue. In breast cancer the under expression of CRBP Citation15-17 has been considered to be due to the altered metabolism of retinol. In contrast with this statement, in several examples of carcinogen-induced tumors as well as in some “spontaneous” ones, it was observed that the level of CRBP(s) (CRABP and CRBP) in the tumor was considerably higher than in the adjacent normal tissue [Citation18,Citation19].
As reported in our previous papers, bovine milk xanthine oxidase (XO), the O2-dependent form of the Xanthine oxidoreductase system (XOR), is capable of oxidizing all-trans-retinol (t-ROL) bound to CRBP (holo-CRBP) to all-trans-retinaldehyde (t-RAL), and all-trans- and 9-cis-retinaldehyde to all-trans- and 9-cis-retinoic acid [Citation10,Citation20]. Given that, a significant amount of xanthine dehydrogenase (XDH), the NAD+-dependent form of XOR, is expressed in the normal human mammary epithelial cells at confluence [Citation21,Citation22], we have evaluated in the HMEC cell line the possible role of XDH in retinoic acid biosynthesis, starting from t-ROL. The participation of CRBP(s) in this process has been also evaluated. Here, we report that NAD+-dependent XDH, cooperated by CRBPs and CRABPs, is responsible for all-trans-retinoic acid (t-RA) biosynthesis and we suggest a novel oxidation pathway of t-ROL to t-RA.
Experimental procedures
Chemicals
All chemicals were from Sigma-Aldrich (Milan, Italy) except NaCl, sucrose, hydroxylamine-HCl, ammonium and sodium acetate and potassium phosphate that were from Merck Group (Milan, Italy), and HPLC-grade acetonitrile, ethanol, 1,4-dioxane, hexane and dichloromethane that were from J.T. Baker. Centriprep, Centricon (YM-50 and YM-10 membranes) and Millex-FG13 filter (0.22 μm pore size), were furnished by Millipore. [11,12-3H]-trans-retinol ([3H]-trans-retinol) and [11,12-3H]-trans-retinoic acid ([3H]-trans-retinoic acid) were from NEN Life Science Research Products, Perkin Elmer. Scintillation cocktails were furnished by Canberra Packard.
Instruments
The Gilson analytical liquid chromatograph used through-out this work consisted of two Model 306 pumps, one 811B dynamic mixer, a Model 234 auto injector equipped with a 100 or 20 μl injection loop, a 122 fluorometric detector, a Waters 2487 dual λ absorbance detector, a radiomatic FLO-ONE βeta 500 TR Series Flow Scintillation Analyzer furnished by Packard a Canberra Company, and Unipoint LC System Software (v. 3.3) (Gilson Italia, Milan, Italy), or a Flo-One/beta F1B IC program (Radiomatic, Tampa, FL) for acquisition and elaboration of data. The Automated Micro plate Reader Lambda E for immunoassay was furnished by Bio-Tek Instruments-Coordination Center Europe (France).
Cells and cell culture
Human mammary epithelial cells (HMEC) were obtained from Clonetics (San Diego, CA). Cells were maintained in mammary epithelial basal medium (MEBM) supplemented with 0.05% bovine pituitary extract and growth factors (MEGM) in a humidified environment at 37°C with 5% CO2 in air. All experiments were performed using HMECs between passages 6-11. Cells were collected when about 90% of confluence was reached.
Isolation and identification of xanthine dehydrogenase from mammary epithelial cells
Homogenate was prepared by cavitation of a cell suspension (about 30 × 106 cells in 3 mL of 0.25 M sucrose, 20 mM Tris-HCl pH 7.0, 0.4 mM EDTA), with or without 1 mM glutathione (GSH) in a pre-cooled cell disruption bomb (Parr Instrument Company) at a nitrogen pressure of 600 psi for 10 min. The homogenate was centrifuged at 110,000 × g for 60 min in a L8-M Beckman ultracentrifuge and 3 mL of supernatant corresponding to about 24 × 106 cells were gel filtered on Shodex KW 804 column (Waters) injecting cytosol aliquots of 100 μl (74 μg protein) to gain separation of XDH, retinoid binding proteins (CRBP(s)) and ADH. Protein content of cytosol was determined using the Sigma 690-A kit. Proteins, eluted with 50 mM Tris-HCl pH 7.4 at a flow rate of 1 mL/min, were monitored measuring their fluorescence (ex. 350 nm/ em. 480 nm) and absorbance at 280 nm and at 350 nm to identify XDH and CRBP(s); ADH was collected in the elution range of bovine serum albumin (66 kDa). Fractions were collected, pooled and concentrated to a third of their initial volume (about 1 mL) with Centriprep YM-50 membrane (Fraction I) and Centripep YM-10 membrane (Fraction II and III). The pooled fraction containing XDH (Fraction I) was added of 1 mM glutathione (GSH) before concentration with Centripep and after flushing with N2 was saved at -80°C. The further purification and analysis of CRBP(s) fraction from human mammary epithelial cells were performed as reported in reference [Citation10].
Xanthine dehydrogenase detection on western blot by chemiluminescence
HMEC cytosol, horse liver ADH (Sigma), buttermilk XO (Sigma X-4500, grade III), and purified fractions were analyzed on an 8% SDS-polyacrylamide gel with broad range prestained standard proteins (Bio-Rad). Gel was stained with red ponceau and blotted to nitrocellulose. In the same run, prestained standard proteins and XDH (Fraction I) after electro migration were directly blotted to nitrocellulose and blocked for 1 h with nonfat dry milk in TBS containing 0.05% Tween 20 and subsequently incubated for 3 h with Abs specific to ADH (Ab, mouse IgM; United States Biological, DBA Italia), and XO (Ab, mouse IgM; United States Biological, DBA Italia). After washing, the blot was incubated for 1 h with horseradish peroxidase-conjugated anti mouse Ab (Amersham) and visualized using an enhanced chemiluminescence detection system (SuperSignal West Dura Extended duration Substrate; Pierce Chemical Co., Rockford, IL).
Xanthine dehydrogenase immunoassay detection
The XDH content in cytosol and purified preparations from HMEC cells was evaluated, employing Mab and Pab antibodies, by a sandwich ELISA immunoassay. Briefly, a polystyrene flat bottom well plate (IWAKI brand SciTech Division, Asahi Techno Glass, Japan) coated with IgM mouse anti-human xanthine oxidase (Mab) (United States Biological; Catalog No X0980-05X, DBA Italia) was incubated overnight at 4° C, rinsed afterwards with 50 mM phosphate buffered saline, pH 8.0, containing 0.05% Tween 20 (PBS/T). The remaining protein binding sites were blocked with 1% BSA in PBS/T incubating for 1 h at 4°C. Then antigen samples to test (50–100 μL) were added and incubated for 2 h at 4°C. Unbound antigen material was removed washing four times with PBS/T. Then rabbit anti-bovine XO/AO (Pab) (United States Biological; Catalog No X0980-20, DBA Italia) conjugated with horseradish peroxidase, at dilution 1:20 000, was added and the plate incubated for 2 h at 4°C. Unbound Pab anti XO/AO was removed washing four times with PBS/T and the peroxidase substrate was added. After 30 min incubation at room temperature the micro plate was read at 450 nm. A standard curve of xanthine oxidase was constructed, in the concentration range of 80–640 ng, to compare the absorbance measured for the samples.
Enzyme assays
Retinoid solutions were performed as previously reported [Citation10].
Homogenate and cytosol assays with free t-ROL or t-RAL
100 μL of homogenate or cytosol were incubated with 25 μL of 2 mM NAD+, 25 μL of a 0.25–2 μM t-ROL or t-RAL in 0.12 mM Tween 80 and 100 μL of 50 mM Tris-HCl, pH 7.4, in a final volume of 250 μL. After 10 min of incubation at 37° C, the mixture was acidified by adding 20 μL of 0.5 M acetic acid, and retinoids were extracted by adding 3 mL of hexane, shacking for 1 min in the dark. The hexane phase was dried under nitrogen and saved at − 20°C. One third of the sample was analyzed as reported in reference [Citation23].
CRBP(s)-retinol mixture preparation
50 μL of t-ROL solutions (0.125, 0.25, 0.5, 1 and 2μM) in 0.12 mM Tween 80 and 50 mM Tris-HCl, pH 7.4 were combined with 200 μL of CRBP(s) fraction and kept at 22°C a few minutes before the assay.
XDH assays with free and bound t-ROL
200 μL of XDH preparation (3.4 μg proteins) were incubated with 50 μL of 2 mM NAD+ and 50 μL of 0.25-2 μM t-ROL in 0.12 mM Tween 80, or 250 μL of t-ROL-CRBP(s) mixture, in a final volume of 500 μL. After 10 min of incubation at 37° C, the sample was treated as above reported.
XDH assays in presence of oxypurinol
For the study on the inhibitory effect by purine analogs, 0.5–10 μM oxypurinol in 10 μL of 0.005 N NaOH were added to the incubation mixture before the addition of the enzyme protein.
XDH assays with xanthine
200 μL of partially purified XDH was incubated in a medium containing 50 mM Tris-HCl pH 7.4, 400 μM NAD+ and 20 μM xanthine in 0.005 N NaOH, solubilized with the aid of ultrasonic bath in a final volume of 500 μL. After 10 min of incubation at 37°C the mixture was diluted 1:1 (v/v) with cold 47 mM potassium phosphate buffer, pH 4.65 and filtered through Millex-FG13 membranes. 100 μL of sample were analyzed following indications reported in reference [Citation24]. Xanthine and uric acid peaks were detected at 264 nm and 295 nm respectively.
Enzyme assay after immune-precipitation
800 μL of XDH preparation was treated with 60 μL (3 μg) of a diluted solution (1:2000) of rabbit anti-bovine XO/AO (Pab) antibodies, incubated for 2 h at 4°C, centrifuged at 20 000 rpm for 10 min, and filtered through Millex-FG13 membranes. After this, 200 μL aliquots of the treated XDH preparation were incubated with CRBP(s) as above reported (200 nM of t-ROL). The same preparation was assayed with xanthine.
Supplementary enzyme assays
Alcohol dehydrogenase activity
The reaction mixture containing 50 mM Tris-HCL, pH 7.8, ethanol in the concentration range of 0.5–5.0 mM, and β-NAD+ at the concentration range of 200–400 μM, was assayed with the HMEC preparations in a volume of 2.5 mL. Formation of NADH was measured at 25 °C in a Perkin-Elmer 650–10S fluorescence spectrophotometer by following the fluorescence emission at 490 nm with the excitation wavelength at 340 nm, for 5 min at 30 s intervals.
t-RAL oxime determination
XDH (Fraction I) was assayed with 500 nm free or bound t-ROL in 50 mM Tris-HCL pH 7.4, 50 μL of 2 mM NAD+ in a volume medium of 500 μL. After 10 min of incubation at 37° C, the mixture was stopped with a 500 μM hydroxylamine-HCl solution buffered at pH 6.7 with sodium bicarbonate [Citation25]. After dilution of the samples to one mL with water, retinoids were extracted by addition of two volumes of a methanol/dichloromethane mixture (1:1 v/v), vortex mixed for 1 min, allowed to stand for 5 min, and mixed for a further 1 min.
At this stage, 0.8 mL of the lower dichloromethane phase was withdrawn and dried under N2. The dry residue was dissolved in 4% dioxane in hexane and analyzed on a μPorasil column (Waters) by eluting with the same 4% dioxane in hexane [Citation25].
Kinetic data processing
Leatherbarrow Eritacus Software made a provisional estimate of kinetic constants with the aid of the Grafit program. The appropriate velocity equations describing the kinetic behaviors [Citation26] were verified. A nonlinear iterative calculation using the Enzyfit program (Sigma-Aldrich) was also performed to calculate the kinetic parameters.
Statistical analysis
Statistical significance of the data was evaluated using Student's t-test [Citation27] and probability values below 0.05 (P < 0.05) were considered significant. Results are expressed as mean ± S.D. from the indicated set of experiments.
Results
Purification of the protein that oxidizes t-Retinol to t-Retinoic Acid
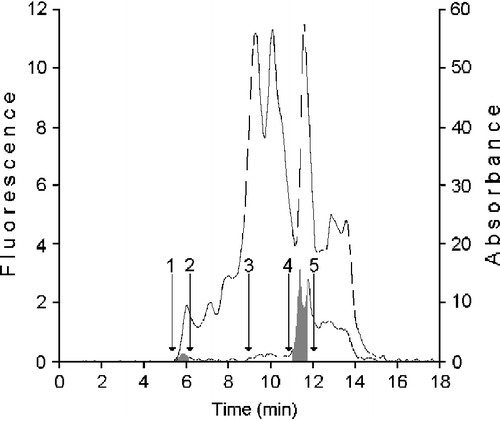
In order to investigate the metabolic system that in the human mammary epithelial cells conduces to retinoic acid synthesis, we have purified from HMEC cytosol the protein catalyzing the t-ROL oxidation, performing a gel permeation chromatography on KW 804 column. Furthermore, considering the possible participation of CRBP(s) in the conversion process of t-ROL to t-RA, we have purified from the same source a fraction containing these specific binding proteins. The fractions collected during the gel chromatography of HMEC cytosol on KW 804 column are shown in . The elution profile reported in this figure shows a first peak, containing XDH (Fraction I), positive for fluorometry (grey area; RT 5.90 ± 0.3), where proteins of about 200-400 kDa are present, and a second peak, it also positive for fluorometry and containing the CRBP(s) (gray area; RT 11.47 ± 0.3), where proteins with masses between 14 and 20 kDa are present (Fraction III). Besides, in the fraction containing proteins with masses between 100-40 kDa (Fraction II; RT 8.5 ± 1.0) ADH enzyme was also collected. A kit of protein markers with masses in the range of 12.4 and 200 kDa, and dextran blue (2,000 kDa) were used for column calibration.
Figure 1 Purification of protein showing retinol dehydrogenase activity and cellular binding proteins from cytosol of human mammary epithelial cells. 100 μL cytosol (1.4 × 106 cells − 37 μg protein) were gel filtered on a KW 804 column and eluted at 1 mL min-1 with 50 mM Tris HCl pH 7.4 containing 1 mM glutathione. Elution peaks were monitored by absorbance at 280 nm (top trace) and fluorescence (bottom trace). Iterative analyses were carried out and fraction peaks were collected from the area containing XDH (Fraction I; RT 5.90 ± 0.3), ADH (Fraction II; RT 8.5 ± 0.3) or cellular retinoid binding proteins (Fraction III; RT 11.47 ± 0.3) (grey peaks). Positions of standards used as molecular weight markers were: 1) blue dextran (2.000 kDa); 2) β-amylase (200 kDa); 3) bovine serum albumin (66 kDa); 4) bovine erythrocytes carbonic anhydrase (29.3 kDa); 5) horse heart cytochrome C (12.4 kDa).
Because the CRBP(s) are characterized by similar masses and given the possibility of the existence of different isoforms in HMEC, it was advisable to further purify and characterize this fraction, performing the procedure already described [Citation10]. Although we evidenced with the aid of radio-chromatography assay different isotypes of CRBPs (CRBP III and CRBP I) Citation28-32 and CRABPs (CRABP II and CRABP I) [Citation33,Citation34], we did not success to perform the separation of each of them (). The amounts of CRBPs and CRABPs available in the sample were determined by measuring the specific binding activity assayed versus the two [3H]-retinoids (). In 50 μl of the analyzed sample containing 2.018 μg of total protein, there were in all about 8.39 ng of CRBPs (about 5.6 pmoles) and 28.6 ng of CRABPs (19.6 pmoles). This CRBP(s) preparation was used to optimise t-ROL oxidation performed by xanthine dehydrogenase.
Figure 2 CRBPs and CRABPs were analyzed and quantified by gel permeation chromatography and radioligand binding activity versus all-trans-retinol or all-trans-retinoic acid. In the Protein Pak I 60 chromatogram of CRBP-3[H]-all-trans-retinol, radioactivity relative to the bound t-ROL was distributed in two peaks (a). Two wider peaks were separated in the CRABP-3[H]-all-trans-retinoic acid chromatogram (b). Grey areas are radioactive peaks; light area is absorbance at 280 nm. Results from a representative of three separate experiments are shown.
![Figure 2 CRBPs and CRABPs were analyzed and quantified by gel permeation chromatography and radioligand binding activity versus all-trans-retinol or all-trans-retinoic acid. In the Protein Pak I 60 chromatogram of CRBP-3[H]-all-trans-retinol, radioactivity relative to the bound t-ROL was distributed in two peaks (a). Two wider peaks were separated in the CRABP-3[H]-all-trans-retinoic acid chromatogram (b). Grey areas are radioactive peaks; light area is absorbance at 280 nm. Results from a representative of three separate experiments are shown.](/cms/asset/546db99a-d5cf-42f2-af6a-859b639ab3cb/ienz_a_258308_f0002_b.gif)
All-trans-Retinoic acid biosynthesis was performed by using free or bound retinoids
Optimal assay conditions were fixed after preliminary assays with t-ROL (500 nM) and all-trans-retinaldehyde (t-RAL) (250 nM) varying the incubation time, the protein aliquot, and the concentration of NAD+ and Tween (data not shown). In routine assays, NAD+ was 200 μM, detergent 0.012 mM, retinoid 200 nM and incubation time 10 minutes. Crude HMEC preparation (homogenate or cytosol) and the chromatography purified one (Fraction I), assayed with 200 nM t-ROL or t-RAL showed higher activity rate for t-RAL with respect to t-ROL (). The activity of the purified enzyme assayed with t-ROL-CRBP(s) was found very high with respect to that assayed with free t-ROL. These results are in agreement with those obtained assaying bovine buttermilk XO [Citation10], for which we determined a higher Kcat value toward holo-CRBP with respect to free t-ROL. The participation of CRABPs in the kinetics of retinoic acid synthesis has also to be taken into account as these can drain the formed t-RA and thus favour t-RA synthesis.
Table I. Retinoic acid biosynthesis rates in human mammary epithelial cells.
Xanthine dehydrogenase preparation does not contain ADH
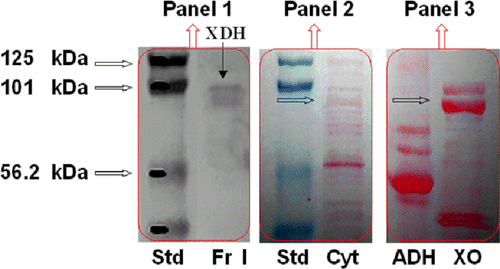
Oxidative activity toward ethanol was determined in the HMEC homogenate and cytosol (0.5 and 1.2 nmoles min− 1 mg− 1), but it was not possible to ascribe it to ADH or XDH [Citation10,Citation35]. However, given the very different mass of the two enzymes (XDH 300 kDa, ADH 80 kDa), they were separated by gel chromatography on KW 804 (), as demonstrated by the highest activity toward ethanol detected in Fraction II (2.65 nmoles min− 1 mg− 1). To test the absence of ADH enzyme in the XDH purified fraction, we have electrophoresed in SDS-PAGE the HMEC cytosol and purified Fraction I, followed by western blotting with Abs specific to ADH and XO, as described in Experimental Procedure. The immunoblot analysis of Fraction I showed the immunofluorescent bands of XDH, while there was no evidence of immunofluorescent bands recognizable as alcohol dehydrogenase in this pattern ().
Figure 3 Western analysis of HMEC cytosol and HPLC purified fractions. SDS-PAGE electropherogram relative to prestained protein standards (Std), HMEC cytosol (Cyt), horse liver ADH (ADH) and buttermilk XO (XO) stained with red Ponceau (Panel 2 and 3), is compared to Western blot of the XDH purified fraction immunostained with XO and ADH antibodies, detected by chemiluminescence (Panel 1). One representative of at least three independent experiments are shown.
Xanthine dehydrogenase performs the straight catalytic conversion of all-trans-retinol to all-trans-retinoic acid
In order to clarify if the catalytic behavior of XDH in t-ROL to t-RA oxidation is comparable to that of XO, for which the demonstrated sequence of oxidation is t-ROL → t-RAL → t-RA [Citation10], and then establish if t-RAL is a free intermediate product in the t-ROL → t-RA conversion catalyzed by XDH, at the end of the incubation time, samples were treated with hydroxylamine to evaluate the presence, in the incubation medium, of t-RAL oxime derivatives (see t-RAL oxime determination in Supplementary Enzyme Assay). The missed formation of t-RAL is strongly indicative of the XDH ability to directly oxidize t-ROL to t-RA. Moreover, the absence of t-RAL in the incubation medium as product of t-ROL oxidation, confirms the data reported by some authors [Citation14].
Kinetic properties of xanthine dehydrogenase toward All-trans-retinol
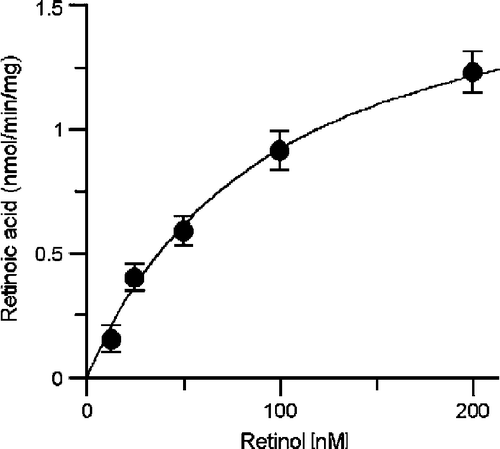
Kinetic constants for XDH catalyzing t-ROL oxidation were determined, performing two independent assays, by varying t-ROL concentration (12.5–200 nM), in the presence of fixed amount of CRBPs (44.8 nM), CRABPs (156.8 nM), and XDH (3.3 nM). The initial kinetic parameter values were estimated using the Eadie-Hofstee plot. The average Km value was 103 ± 9.34 nM ( ± S.E.) and the average Vmax value was 1.88 ± 0.071 nmol/min/mg ().
Figure 4 Km value for retinol determined by XDH kinetics in the presence CRBP is of nanomolar order. t-RA synthesis rates were determined assaying XDH purified from HMEC cells (3.3 nM) with increasing amounts of t-ROL (12.5-200 nM) in the presence of fixed apo-CRBP (44.8 nM) and apo-CRABP (156.8 nM) concentration. Kinetic data represent mean ± S.D. of triplicates from a representative experiments.
Retinol dehydrogenase activity of xanthine dehydrogenase was inhibited by oxypurinol
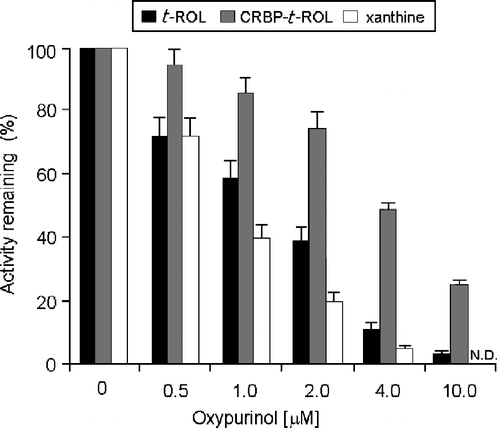
Xanthine dehydrogenase was found to be well expressed in confluent HMEC cells and when assayed with xanthine its activity was full inhibited by 4 μM oxypurinol (). A similar inhibitory effect was also evidenced on the enzyme when operative on t-ROL; on the other hand, the inhibitory effect of 4 μM oxypurinol tested on the oxidative reaction of t-ROL-CRBP resulted to be only 48%. In this latter case, the lesser sensitivity to the inhibitor shown by the enzyme is probably due to the influence of the CRBPs and was overcome at higher oxypurinol concentrations (10 μM).
Figure 5 Comparison of the inhibitory effects performed by oxypurinol in the oxidation of xanthine, all-trans-retinaldehyde or all-trans-retinol in human mammary epithelial cells. 10 μM xanthine, 200 nM free t-ROL or 200 nm t-ROL bound to CRBP(s), were incubated with the xanthine dehydrogenase purified from HMEC cytosol at oxypurinol concentration of 0.5- 10 μM. The inhibitory effect of oxypurinol against xanthine was already distinguished at the lowest concentration (0.5 μM) (P < 0.05 vs. control) and it was fully realized at 4 μM concentration (P < 0.01 vs. control). The inhibitory effect observed against t-ROL bound to CRBP(s) is instead accomplished at a higher oxypurinol concentration (10 μM) (P < 0.01 vs. control). Results from a representative of three separate experiments are shown, reported as the mean ± S.E.M of triplicate determinations. Statistical significance, calculated with Student's paired t-test, refers to a comparison of sample incubated with or without oxypurinol.
Another finding that supports the equivalence between retinol dehydrogenase and xanthine dehydrogenase was the full loss of catalytic activity toward t-ROL and xanthine of the purified enzyme fraction, after treatment with rabbit anti-bovine XO/AO (Pab) antibodies. XDH content was evaluated in cytosol and purified preparations from HMEC cells employing mouse anti-human xanthine oxidase (Mab) and rabbit anti-bovine XO/AO (Pab) antibodies. A significant amount of the enzyme protein (about 300 ng) was detected assaying cytosol aliquots from 250 × 103 HMEC. The XDH amount recovered in the purified fraction was about 83% with respect to the crude preparation ().
Table II. Detection and content of xanthine dehydrogenase in human mammary epithelial cells.
Xanthine dehydrogenase native status is necessary to maintain its retinol dehydrogenase activity
The purification procedure of XDH and the measurement of its catalytic activity versus t-ROL and t-RAL were performed in presence of 1 mM GSH to prevent the conversion of the dehydrogenase into the form incapable to oxidize t-ROL to t-RA, yet keeping its catalytic activity versus t-RAL and xanthine. Cytosol obtained from a cell homogenate prepared without glutathione, almost devoid of activity versus t-ROL, showing only 20% activity with respect to the treated homogenate, was able to partly recover it (40%) after a few hours treatment with 2 mM glutathione at 4°C (data not shown).
Discussion
Several studies report on the inhibitory effect of the retinoic acids in growth of mammary tumors and breast cancer cells in vitro and on its action, mediated by the specific classes of receptors (RAR and RXR) Citation36-38. Experimental findings of a retinoid metabolism, dedicated to the t-RA production, obtained with indirect rather than with direct observations, comprise detection and estimation of CRABP [Citation32], or [3H]-t-ROL processing in cultures of normal and cancer mammary epithelial cells [17].
all-trans-Retinol-CRBP is the physiological substrate in the all-trans-Retinoic acid synthesis catalyzed by xanthine dehydrogenase
Three cellular retinol-binding proteins (CRBP I, II and III) with distinct tissue distribution and retinoid-binding properties have been recognized thus far in mammal tissues [Citation31,Citation32]. A number of in in vitro studies indicates that CRBP II and I might support retinol metabolism by facilitating enzymatic reactions involved in retinol storage and utilization, while for CRBP III a possible compensatory role has been hypothesized. However, the precise functional roles of these binding proteins have not been yet defined.
In the present work we report data on the action of a CRBP(s)-supported catalytic system that in human mammary epithelial cells directly oxidizes t-ROL to t-RA with the initial velocity of 1.223 nmol * min− 1 * mg− 1. The initial velocity at the same t-ROL concentration, but in absence of the CRBP(s) fraction, was only 0.080 nmol * min− 1 * mg− 1, giving clear indication on the supporting role of CRBP(s) in the reaction. The enzyme was subsequently identified as xanthine dehydrogenase by immunoassay, by its ability to oxidize xanthine to uric acid and its sensitivity to the oxypurinol inhibitory effect. Furthermore, it was also observed that both retinol and xanthine dehydrogenase activities were undetectable after polyclonal XO/AO antibody treatment.
Modification of xanthine dehydrogenase and loss of its catalytic activity toward all-trans-Retinol
It has been well reported that the basis of the conversion of XDH to XO is the oxidation of cysteine thiols to form disulfide bonds Citation39-41, associated with conformational change at the flavin-binding site [Citation42,Citation43], with loss of NAD+ binding affinity [Citation44], and alterations in redox and kinetic properties [Citation45,Citation46]. These structural modifications of XDH, that involve its reversible conversion to the oxidase form, become irreversible when proteolytic cleavage of the protein occurs [Citation47]. The fact that GSH addition to HMEC preparations safeguards the enzyme activity toward t-ROL while untreated preparations became unable to oxidize it, although saving the capacity of oxidizing t-RAL, and the demonstration that buttermilk XO, treated with Na2S, is capable to oxidize t-ROL [Citation10], are both arguments to suggest that the loss of activity toward t-ROL is not ascribable to alterations of the above mentioned protein structure. On the other hand, it has also reported that the greater proportion of inactive XDH and XO is made up of “desulfo” enzyme Citation48-50, in which the molybdenum atom is covalently double-bonded to oxygen, rather than sulfur (Mo = S → Mo = O). We reckon that substitution of sulfur at the molybdenum center of the enzyme caused by reactive oxygen species may be prevented by GSH, either in the XDH or XO forms. Thus, the reconstitution of the native structural organization of the molybdenum centre of XDH and XO desulfo forms [Citation51] is fundamental for the XOR system to express activity toward alcohols. Moreover, in the XDH, the preservation of the native structural organization of both protein and molybdenum centre is the necessary condition to perform the straight oxidation of t-ROL to t-RA.
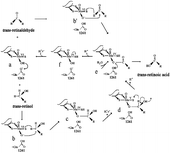
A hypothetic reaction mechanism of xanthine dehydrogenase in all-trans-Retinol and all-trans-Retinaldehyde Oxidation
In the present work we demonstrate the presence, in mammary cells, of a single pathway that goes directly from t-ROL to t-RA. The direct conversion of t-ROL to t-RA was already observed for the soluble enzyme activity from calf liver cytosol [Citation14]. Our findings on t-ROL metabolic pathway addressed by XDH in mammary epithelial cells did not evidence any release of t-RAL as intermediate product; however, when t-RAL is the substrate, it is turned over to t-RA more actively. Such different behavior of XDH versus the two substrates may be counted to the different reactivity status of the C-15 position of t-ROL and t-RAL molecule. It has long been known that the active site of the enzyme possesses a catalytically labile oxygen (Mo-OH group) that represents the proximal oxygen donor to substrate in the course of reaction [Citation52]. On this basis we hypothesize a reaction mechanism of hydroxylation/oxidation of t-ROL, and of t-RAL that is shown in .
Figure 6 Hypothetical scheme of reaction mechanism devised by xanthine dehydrogenase in oxidizing trans-retinol to trans-retinoic acid. A catalytically labile Mo-OH group of the oxidized (MoVI) enzyme (a) initiates catalysis by nucleophililic attack on the carbon center of t-retinol or t-retinaldehyde to be hydroxylated, with concomitant hydride transfer (b and b’). This yields a reduced Mo(IV)-SH that derives from the Mo(VI) = S of the oxidized enzyme (c and e), with the product remaining coordinated to the molybdenum via the introduced hydroxyl group. In the case of t-retinol, after one-electron oxidation a new intermediate (d), so-called (46) “very rapid” Mo(V) species, is formed, and after rearrangement of the intermediate at C-15 a new hydride transfer is necessary (d). This yields a new reduced Mo(IV)-SH (e) that derived from the Mo(V) = S of the enzyme (d). The Mo(IV) intermediate (f) breaks down by direct hydroxide displacement of the product in presence of water.
Wide XDH involvement in retinoid homeostasis
It seems surprising that the xanthine oxidoreductase system (XDH/XO) [Citation44], is able to perform retinol oxidation, since these enzymes are actually well-known for their role in nucleotide adenylate and purine degradation.
The ascertained promising action of retinoic acid and its analogues on the control of cell differentiation and growth in various normal and tumor cells, included mammary epithelial cells Citation53-56, encourages us to extend our investigation to the availability and catalytic efficiency of XDH in cancer breast cells.
In addition, XDH implication in retinoid homeostasis may be also considered to pursue other aims; the conversion of t-ROL to t-RA may in fact prove useful to obtain a prompt decrease of t-ROL level in the cell. Not less useful will prove the contribution of XDH in converting retinaldehyde to retinoic acid thus preventing its accumulation, when formed by carotene cleavage in some tissues [Citation57]. Involvement of XDH in the t-RA biosynthesis in mammary epithelial cells also provides a confirmation for the initial observations on the ability of bovine milk XO to process t-RAL [Citation20,Citation57,Citation58] and t-ROL [Citation10]. In addition, the participation of CRBP(s) in XDH catalysis induces speculation on possible models of molecular interaction between the XDH and retinoid binding proteins, for which models of functional architectures are already available [Citation59,Citation60].
Significance
As a conclusion, this study points to the demonstration that the enzyme already known as xanthine dehydrogenase catalyzes the retinol oxidation; it also states the role of the cellular retinoid binding proteins in supporting its retinol dehydrogenase activity. The process of retinoic acid biosynthesis by XDH is biologically advantageous because it is accomplished in one cellular compartment, the cytoplasm, where both enzyme and retinoid binding proteins are available. Moreover, xanthine dehydrogenase, with the composite organization of its redox centers, at the head of which there is molybdenum, results to be particularly effective when compared with the other enzyme systems involved in retinoic acid synthesis, because unlike these, it seems to perform the conversion of t-ROL into t-RA in a single catalytic step.
In the near future, this study can be useful basis to explain the impaired retinoid metabolism described for tumor human mammary epithelial cells.
Acknowledgements
We wish to thank prof. Luigi Castagnetta and dr. Giuseppe Carruba for providing generous access to the Laboratory of Experimental Oncology Unit, Department of Clinical Oncology, ARNAS-Civico, Palermo. We also thank dr. Letizia Cocciadiferro for help with the cell culture, dr. Grazia M. Granata for radio-chromatography analysis and dr. Monica Zerilli (Department of Surgical and Oncological Sciences, School of Medicine, University of Palermo) for Western analysis. This work was supported by a MURST grant.
References
- Gudas LJ, Sporn MB, Roberts AB. Cellular biology and biochemistry of the retinoids. The retinoids: Biology, chemistry, and medicine, MB Sporn, AB Roberts, DS Goodman. Raven Press, Ltd., New York 1994; 443–520
- Lotan R. Effect of vitamin A and its analogs (retinoid) on normal and neoplastic cells. Biochim Biophys Acta 1980; 605: 33–91
- Orphanos CE. Oral retinoids-present status. Br J Dermatol 1980; 103: 473–481
- Chytil F. Retinoic acid, biochemistry, pharmacology, toxicology and therapeutic use. Pharmacol Rev 1984; 36: 93S–100S
- Kraemer KH, Di Giovanna JJ, Moshell AN, Tarone RE, Peck GL. Prevention of skin cancer in xeroderma pigmentosum with the use of oral isotretinoin. N Engl J Med 1988; 318: 1633–1637
- Kim CL, Leo MA, Lieber CS. Retinol forms retinoic acid via retinal. Arch Biochem Biophys 1992; 294: 388–393
- Blaner WS, Olson JA. Retinol and retinoic acid metabolism. The retinoids: Biology, chemistry, and medicine, B Sporn, AB Roberts, DS Goodman. Raven Press, Ltd., New York 1994; 229–255
- Napoli JL. Retinoic acid biosynthesis and metabolism. FASEB J 1996; 10: 993–1001
- Napoli JL. 17β-Hydroxysteroid dehydrogenase type 9 and other short-chain dehydrogenases/reductases that catalyze retinoid, 17β- and 3α-hydroxysteroid metabolism. Mol Cell Endocrinol 2001; 171: 103–109
- Taibi G, Nicotra CMA. Xanthine oxidase catalyzes the oxidation of retinol. J Enz Inhib Med Chem 2007; 22: 471–476, (240758(07/1122)
- Chen H, Howald WN, Juchau MR. Biosynthesis of all trans-retinoic acid from all trans-retinol: catalysis of all trans-retinol oxidation by human P-450 cytocromes. Drug Metab Dispos 2000; 28: 315–322
- Rexer BN, Zheng WL, Ong DE. Retinoic acid biosynthesis by normal human breast epithelium is via aldehyde dehydrogenase 6, absent in MCF7 cells. Cancer Res 2001; 61: 7065–7070
- Ong DE, MacDonald PN, Gubitosi AM. Esterification of retinol in rat liver: Possible participation by cellular retinol-binding protein and cellular retinol binding protein (II). J Biol Chem 1988; 263: 5789–5796
- Ottonello S, Scita G, Mantovani G, Cavazzini D, Rossi GL. Retinol bound to cellular retinol-binding protein is a substrate for cytosplasmic retinoic acid synthesis. J Biol Chem 1993; 268: 27133–27142
- Jing Y, Zhang J, Bleiweiss IJ, Waxman S, Zelent A, Mira-Y-Lopez R. Defective expression of cellular retinol binding protein type I and retinoic acid receptors α2, β2 and γ2 in human breast cancer cells. FASEB J 1996; 10: 1064–1070
- Kuppumbatti YS, Bleiweiss IJ, Mandeli JP, Waxman S, Mira-Y-Lopez R. Cellular retinol-binding protein expression and breast cancer. J Natl Cancer Inst 2000; 92: 475–480
- Hayden LJ, Satre MA. Alterations in cellular retinol metabolism contribute to differential retinoid responsiveness in normal human mammary epithelial cells versus breast cancer cells. Breast Cancer Res Tr 2002; 72: 95–105
- Chytil F, Ong DE. Mediation of retinoic acid-induced growth and antitumor activity Nature 1976; 260: 49–51
- Ong DE, Markert C, Chiu JF. Cellular binding proteins for vitamin A in colorectal adenocarcinoma of rat. Cancer Res 1978; 38: 4422–4426
- Taibi G, Paganini A, Gueli MC, Ampola F, Nicotra CMA. Xanthine oxidase catalyzes the synthesis of retinoic acid. J Enz Inhib 2001; 16: 275–285
- Kurosaki M, Zanotta S, Li Calzi M, Garattini E, Terao M. Expression of xanthine oxidoreductase in mouse mammary epithelium during pregnancy and lactation: regulation of gene expression by glucocorticoids and prolactin. Biochem J 1996; 319: 801–810
- McManaman JL, Hanson L, Neville MC, Wright MR. Lactogenic hormones regulate xanthine oxidoreductase and β-casein levels in mammary epithelial cells by distinct mechanisms. Arch Biochem Biophys 2000; 373: 318–327
- Frolik CA, Tavela TE, Peck GL, Sporn MB. High-Pressure Liquid Chromatographic determination of 13-cis-retinoic acid and all-trans-retinoic acid in human plasma. Anal Biochem 1978; 86: 743–750
- Kock R, Delvoux B, Greiting H. A comparative study of the concentrations of hypoxanthine, xanthine, uric acid and allantoin in the peripheral blood of normals and patients with acute myocardial infarction and other ischaemic diseases. Eur J Clin Chem Clin Biochem 1993; 31: 303–310
- Groenendijk GWT, Jensen PAA, Bonting SL, Daemen FJM. Analysis of geometrically isomeric vitamin A compounds. Meth Enzymol 1980; 67: 203–220
- Segel IH. Enzyme kinetics. Wiley Interscience, New York 1975
- Snedecor G, Cochran W. Statistical Methods. Iowa State University Press, Ames, IA 1967; 120–134
- Erikson U, Das K, Busch C, Nordlinder H, Rask L, Sundelin J, Säilström J, Peterson PA. Cellular retinol-binding protein quantitation and distribution. J Biol Chem 1984; 259: 13464–13470
- Levin MS, Li E, Ong DE, Gordon JI. Comparison of the tissue-specific expression and developmental regulation of two closely linked rodent genes encoding cytosolic retinol-binding proteins. J Biol Chem 1987; 262: 718–7124
- Schaefer WH, Kakkad B, Crow JA, Blair IA, Ong DE. Purification, primary structure characterization, and cellular distribution of two form of cellular retinol-binding protein, type II from adult rat small intestine. J Biol Chem 1989; 264: 4212–4221
- Thompson JR, Bratt JM, Banaszak LJ. Crystal structure of cellular retinoic acid binding protein I shows increased access to the binding cavity due to formation of an intermolecular β-sheet. J Mol Biol 1995; 252: 433–446
- Piantedosi R, Ghyselinck N, Blaner WS, Vogel S. Cellular retinol-binding protein type III is needed for retinoid incorporation into milk. J Biol Chem 2005; 280: 24286–24292
- Fiorella PD, Giguère V, Napoli JL. Expression of Cellular Retinoic Acid-binding Protein (Type II) in Escherichia Coli. J Biol Chem 1993; 268: 21545–21552
- Folli C, Calderone V, Ottonello S, Bolchi A, Zanotti G, Stoppini M, Berni R. Identification, retinoid binding, and x-ray analysis of a human retinol-binding protein. Proc Nat Acad Sci USA 2001; 98: 3710–3715
- Castro GD, Delgado de Layño AMA, Costantini MH, Castro JA. Cytosolic xanthine oxidoreductase mediated bioactivation of ethanol to acetaldehyde and free radicals in rat breast tissue. Its potential role in alcohol-promoted mammary cancer. Toxicology 2001; 160: 11–18
- Mehta RG, Moon RC. Hormonal regulation of retinoic acid binding proteins in the mammary gland. Biochem J 1981; 200: 591–595
- Roman SD, Clarke CL, Hall RE, Alexander IE, Sutherland RL. Expression and regulation of retinoic acid receptors in human breast cancer cells. Cancer Res 1992; 52: 2236–2242
- Budhu A, Gillilan R, Noy N. Localizzation of the RAR interaction domain of cellular retinoic acid binding protein-II. J Mol Biol 2001; 305: 939–949
- Stirpe F, Della Corte E. The regulation of rat liver xanthine oxidase. Conversion in vitro of the enzyme activity from dehydrogenase (type D) to oxidase (type O). J Biol Chem 1969; 244: 3855–3863
- Waud WR, Rajagopalan KV. The mechanism of conversion of rat liver xanthine dehydrogenase from an NAD-dependent form (type D) to an O2-dependent type (type O). Arch Biochem Biophys 1976; 172: 354–364
- McManaman JL, Bain DL. Structural and conformational analysis of the oxidase to dehydrogenase conversion of xanthine oxidoreductase. J Biol Chem 2002; 277: 21261–21268
- Massey V, Schopfer LM, Nishino T, Nishino T. Differences in protein structure of xanthine dehydrogenase and xanthine oxidase revealed by reconstitution with flavine active site probes. J Biol Chem 1989; 264: 10567–10573
- Saito T, Nishino T, Massey V. Differences in environment of FAD between NAD-dependent and O2-dependent types of rat liver xanthine dehydrogenase shown by active site probe study. J Biol Chem 1989; 264: 15930–15935
- Hille R, Nishino T. Flavoprotein structure and mechanism. 4. Xanthine oxidase and xanthine dehydrogenase. FASEB J 1995; 9: 995–1003
- Saito T, Nishino T. Differences in redox and kinetic properties between NAD-dependent and O2-dependent types of rat liver xanthine dehydrogenase. J Biol Chem 1989; 264: 10015–10022
- Hunt J, Massey V. Purification and properties of milk xanthine dehydrogenase. J Biol Chem 1992; 267: 21479–21485
- Kooij A, Schiller HJ, Schijns M, et al. Conversion of xanthine dehydrogenase into xanthine oxidase in rat liver and plasma at the onset of reperfusion after ischemia. Hepatology 1994; 19: 1488–1495
- Bray RC. The enzymes3rd edn, PD Boyer. Academic Press, New York 1975; Vol XII: 299–419
- Ikegami T, Nishino T. The presence of desulfo xanthine dehydrogenase in purified and crude enzyme preparations from rat liver. Arch Biochem Biophys 1986; 247: 254–260
- Abadeh S, Killacky J, Benboubetra M, Harrison R. Purification and partial characterization of xanthine oxidase from human milk. Biochim Biophys Acta 1992; 1117: 25–32
- Massey V, Brumby PE, Komai H, Palmer G. Studies on milk xanthine oxidase. Some spectral and kinetic properties. J Biol Chem 1969; 244: 1682–1691
- Okamoto K, Matsumoto K, Hille R, Eger BT, Pai EF, Nishino T. The crystal structure of Xanthine oxidoreductase during catalysis: Implications for reaction mechanism and enzyme inhibition. Proc Natl Acad Sci USA 2004; 101: 7931–7936
- Shao ZM, Dawson ML, Li XS, Rishi AK, Sheikh MS, Han QX, Ordonez JV, Shroot B, Fontana JA. p53 independent G0/G1 arrest and apoptosis induced by a novel retinoid in human breast cancer cells. Oncogene 1995; 11: 493–504
- Seewaldt VL, Kim JH, Caldwell LE, Johnson BS, Swisshelm K, Collins SJ. All-trans-retinoic acid mediates G1 arrest but no apoptosis of normal human mammary epithelial cells. Cell Growth Differ 1997; 8: 631–641
- Farhana L, Dawson M, Rishi AK, Zang Y, Van Buren E, Trivedi C, Reichert U, Fang G, Kirschner MW, Fontana JA. Cyclin B and E2F-1 expression in prostate carcinoma cells treated with the novel retinoid CD437 are regulated by the ubiquitin-mediated pathway. Cancer Res 2002; 62: 3842–3849
- Rishi AK, Zhang L, Boyanapalli M, Wali A, Mohammad RM, Yu Y, Fontana JA, Hatfield JS, Dawson MI, Majumdar APN, Reichert U. Identification and characterization of a cell cycle and apoptosis regulatory protein-1 as a novel mediator of apoptosis signaling by retinoid CD437. J Biol Chem 2003; 278: 33422–33435
- Futterman S. Enzymatic oxidation of vitamin A aldehyde to vitamin A acid. J Biol Chem 1962; 237: 677–680
- Olson JA. The metabolism of vitamin A. Pharmacol Rev 1967; 19: 559–596
- Franzoni L, Lucke C, Pérez C, Cavazzini D, Rademacher M, Ludwig C, Spisni A, Rossi GL, Ruterjans H. Structure and backbone dynamics of apo- and holo-cellular binding protein in solution. J Biol Chem 2002; 277: 21983–21997
- Lu J, Cistola DP, Li E. Two homologous rat cellular retinol-binding proteins differ in local conformational flexibility. J Mol Biol 2003; 330: 799–812